Week 7: unusually-dividing unicellular organisms
Shibani: Chromochloris zofingiensis (Chlorophyceae) Divides by Consecutive Multiple Fission Cell-Cycle under Batch and Continuous Cultivation
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7920477/ – conduct consecutive DNA synthesis and divisions of the nucleus to produce 8 or 16 nuclei before it divides into 8 or 16 daughter cells, respectively.
https://academic.oup.com/jxb/article/65/10/2585/573549 – green algae – 4-16 daughter cells stably – Light and temperature affect cell-cycle progression
Kazeem: dinoflagellate mutant with higher frequency of multiple fission. https://link.springer.com/article/10.1007/BF02680134 Crypthecodinium cohnii Biecheler propagates by both binary and multiple fission. By a newly developed mutagenesis protocol based on using ethyl methanesulfonate and a cell size screening method, a cell cycle mutant,mƒ2, was isolated with giant cells which predominantly divide by multiple fission. – also Plasmodium does this
Jessica: https://www.nature.com/articles/nrmicro1096 – in bacteria. Multiple different division strategies!
Kostas: algal paper from above esp Scenedesmus – with a different mtDNA code! https://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=tgencodes#thetop Not just Plasmodium – other unicell parasites can do both binary and multiple, depending on ecological and environmental pressures. In algae, choice 4-8-16 depends on predator avoidance.
Rajneesh: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC88984/ Giardia – binary division? Longitudinal binary fission https://www.cdc.gov/parasites/giardia/pathogen.html#:~:text=Giardia%20trophozoites%20multiply%20by%20splitting,lining%20of%20the%20small%20intestine – MROs not mitochondria.
Iain: HeLa (to be populated)
Week 5: marine microalgae (maybe with fragmented mtDNA)
Olav: algal blooms in fish farms: Pseudochattenella. In Norway, fish farms often in small fjords – lots of waste concentrated – lots of algal blooms. Chile – global competitor for salmon, suffered from this problem, now no longer a big player. https://www.sciencedirect.com/science/article/pii/S1568988316302116 quite a lot of uncertainty in the mechanism of toxicity – whether live cells, or algal extracts, are required?
Kostas: (quotes from papers) Diplonema papillatum https://link.springer.com/article/10.1186/s12915-023-01563-9
minicircular mitochondrial genomes in red algae https://www.nature.com/articles/s41467-023-39084-2 – including some single celled examples in Fig 1 e.g. a Rhodosorus marinus CCMP1338.
Arguably, the most bizarre mitochondrial DNA (mtDNA) is that of the euglenozoan eukaryote Diplonema papillatum. https://academic.oup.com/nar/article/39/3/979/2409609
[Multiple Independent Changes in Mitochondrial Genome Conformation in Chlamydomonadalean Algae] https://academic.oup.com/gbe/article/9/4/993/3738640
[MITOCHONDRIAL DNA IN THE OOGAMOCHLAMYS CLADE (CHLOROPHYCEAE): https://onlinelibrary.wiley.com/doi/full/10.1111/j.1529-8817.2009.00753.x
“there are several examples of mitochondrial genomes fragmented into circular or linear subgenomic chromosomes with the most extreme and well-documented examples from the ichthyosporean Amoebidium parasiticum (Burger et al. 2003a) and the diplonemid Diplonema papillatum (Marande et al. 2005, Marande and Burger 2007) whose mitochondrial fractions contain >100 distinct DNA molecules, which are linear and circular, respectively.”
Kazeem: Lobochlamys genus https://en.wikipedia.org/wiki/Lobochlamys – fragmented mtDNA – question about linear vs circular vs …? Structures. https://academic.oup.com/mbe/article/25/3/487/1058097 – Polytomella capuana has very high GC content, linear hairpin structure
Rajneesh: Nannochloropsis https://en.wikipedia.org/wiki/Nannochloropsis use in biofuel – high growth rate, high lipid accumulation. Have chlorophyll a but not chlorophyll b or chlorophyll c; 2-5um. Improved N. gaditana productivity by selecting on pigmentation https://biotechnologyforbiofuels.biomedcentral.com/articles/10.1186/s13068-020-01718-8
Week 4 (2024): organisms with mtDNA very different to humans, and why in terms of their lifestyle?
Kazeem- Fruit flies (Drosophila): model organism, short lifespan (60-80 days, females live longer), fully sequenced genome. Genomic structure of mtDNA different from humans, wider nucleotides (A-C). Also transportation of molecules using mitochondrial carriers differs from transportation in humans.
Olav: Giardia lamblia, a parasite. Used to have mitos, but not anymore, also complete lack of mtDNA. They diverged from eukaryotes after they lost their mitos. Why no mt genes? Because they rely on the host for energetic control, can give away all genes.
Shibani: red algae Rhodophyta. Only 10 mt genes. Weird thing: genome into circles and fragmented, each circle has just one gene (one copy number), circles are of different types, circles can also merge and recombine. (We know that ptDNA is much larger, as it has more genes, than mtDNA, potential why in the changing environment)
Maria: seaweed. Dynamics of mitos using a mitotracker, some issues with mitos avoiding them as they might be perceived as toxic. Interesting dynamics in mito movement and division that differ from humans.
Rajneesh: Tardigrade (Iain’s fave), water bear. Model organism for multicellular organism to send to outer space. Mito structure different, to survive in extreme conditions. Much larger mitos (larger genome) in tardigrade, mitos localised in a central region of the cell (in humans they are scattered). Why? Because of the extreme conditions, large mts improve survival and centralised location enhances energy production. Debatable that large mito genome improves survival, examples found of small genomes in organisms in extreme environments.
Belen: Mytilus, common mussel. One more gene for a tRNA than in humans. Many intergenic regions. High frequency of heteroplasmy in males (female and male linages can differ over 20%). Heteroplasmy due to paternal leakage, which is very common in this family. Why? Intertidal environment, higher number of genes and of different types (heteroplasmy) is beneficial for adaptation to the different environmental cycles. Bivalvia in general vary mt genome within this family.
Kostas: Agrostis stolonifera, plant. High mtDNA genome content, hypothesis than ptDNAs have been acquired. 33 protein encoding genes (20 more than humans), 19 tRNAs. 33 times more base pairs than humans. Paper: Complete mitochondrial genome of Agrostis stolonifera: insights into structure, Codon usage, repeats, and RNA editing
Week 40 – ATP budgets
Kostas – four papers
- DNA damage and transcription stress cause ATP-mediated redesign of metabolism and potentiation of anti-oxidant buffering
- Connecting single-cell ATP dynamics to overflow metabolism, cell growth, and the cell cycle in Escherichia coli
- Measuring and modeling energy and power consumption in living microbial cells with a synthetic ATP reporter
- Lacking chloroplasts in guard cells of crumpled leaf attenuates stomatal opening: both guard cell chloroplasts and mesophyll contribute to guard cell ATP levels
2 and 3 particularly good overviews of ATP budgets in single cells
Rajneesh – paper 3. ATP reporter calibrated by reference to bulk measurements of “fixed” ATP conc in a population of bacteria (via luciferase)
Kazeem – ATP requirements in moles (of molecules)
. Suwara, E. Radzikowska-Cieciura, A. Chworos, R. Pawlowska, TheATP-dependent Pathways and Human Diseases, Current MedicinalChemistry 30 (11) (2023) 1232–1255.P. Galichon, M. Lannoy, L. Li, S. Vandermeersch, D. Legouis, M. T.Valerius, J. Hadchouel, J. V. Bonventre, Kidney epithelial proliferationimpairs cell viability via energy depletion.N. Narasimhan, G. Attardi, Specific requirement for ATP at an earlystep of in vitro transcription of human mitochondrial DNA.,Proceedings of the National Academy of Sciences 84 (12) (1987)4078–4082.M. Boiani, V. Gambles, H. Sch ̈oler, Atp levels in clone mouse embryos,Cytogenetic and Genome Research 105 (2-4) (2004) 270–278.J. F. Nascimento, R. O. O. Souza, M. B. Alencar, S. Marsiccobetre,A. M. Murillo, F. S. Damasceno, R. B. M. M. Girard, L. Marchese
L. A. Lu ́evano-Martinez, R. W. Achjian, J. R. Haanstra, P. A. M.Michels, A. M. Silber, How much (atp) does it cost to build atrypanosome? a theoretical study on the quantity of atp needed tomaintain and duplicate a bloodstream-form trypanosoma brucei cell,PLOS Pathogens 19 (7) (2023)
Olav – photoreceptors. ATP requirements – mammals vs insects – backed out through electrochemical observations and modelling https://www.cell.com/current-biology/pdf/S0960-9822(08)01398-5.pdf
Marwa – plant microscopy, wheat epidermis https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6399813/pdf/12918_2019_Article_689.pdf
Shibani – cytoskeletal filaments – 1 ATP per monomer addition (known length). Goldfish epithelial cells. Track motion of cell interface 0.2um/s – calculate how much ATP needed to move this much. 4000 filaments => 4e5 ATP/s cell motion. 2e7 ATP/s protein synthesis.
Iain – quantifying the cell https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4230611/
Feeling for the numbers in biology https://www.pnas.org/doi/10.1073/pnas.0907732106
Week 39 – Tree of Life!
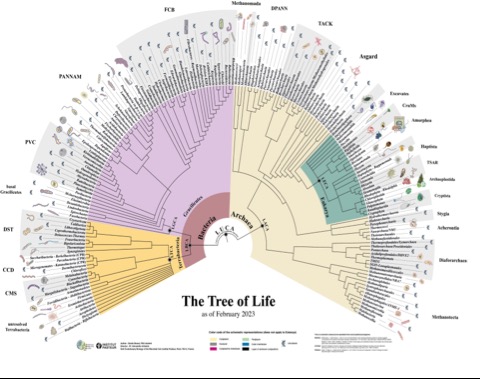
Shibani – Halobacteria – actually archaea, halophiles some with pigments. Some use light to produce ATP, but not with photosynthesis? Purple Halobacterium species owe their color to bacteriorhodopsin, a light-sensitive protein which provides chemical energy for the cell by using sunlight to pump protons out of the cell. https://en.wikipedia.org/wiki/Halobacterium
Kostas – Hodarchaeia – sister clade to eukaryotes. Member of Asgard archaea. Ester-linked lipids on their membrane – usually restricted to bacteria and eukaryotes.
Kazeem – Actinobacteria and Hydrobacteria / Gracilicutes. Terrestrial environment, soil and sediment at high temps. Bioactive molecule production; biocontrol agents. Used for production of tetracycline. Actinomycetota is one of the dominant bacterial phyla and contains one of the largest of bacterial genera, Streptomyces.[7] The Actinomycetota genus Bifidobacterium is the most common bacteria in the microbiome of human infants.[9] Some Siberian or Antarctic Actinomycetota is said to be the oldest living organism on Earth, frozen in permafrost at around half a million years ago.[12][13] https://en.wikipedia.org/wiki/Actinomycetota
Rajneesh – Chloroflexi – https://en.wikipedia.org/wiki/Chloroflexota green non-sulfur bacteria – photosynthetic w unique mechanism. Wide range of terrestrial and aquatic environments.
ATP
Belén: protein synthesis needs ATP, Too much ATP however is unfavorable https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4531837/
Desert species
Olav – Judean date palm – recovered from a 2000 year old seed https://en.wikipedia.org/wiki/Judean_date_palm
Yusra – heat-adapted ants – differences in gene expression in heat stress in ants https://www.biorxiv.org/content/10.1101/2022.11.12.516273v2.abstract
Physiology and genomics of desert plants https://www.sciencedirect.com/science/article/pii/S209012322300125X – CAM metabolism
Rajneesh – black-tailed jackrabbit https://en.wikipedia.org/wiki/Black-tailed_jackrabbit Huge ears! Active at cooler times; adapted to eat hard awkward plants. Classic observational paper from 1976 in Mojave paper https://www.jstor.org/stable/1379705?seq=2
Belén – cyanobacteria? Primary producers in the desert! https://www.bas.ac.uk/data/our-data/publication/cyanobacteria-in-deserts-life-at-the-limit/ “They also accumulate trehalose as a water replacement mechanism to maintain the functional integrity of membranes during anhydrobiosis. Cyanobacteria tolerate high and low extremes of temperature. Their capacity for screening excessive solar radiation (PAR and UVb) by synthesis of “sunscreen” biochemicals whilst retaining a capacity for shade-adaptation. makes them eminently suited for colonisation of diverse lithic habitats. They pioneer the development of microphytic soil crusts which stabilise mobile desert soils. They colonise fissures in rocks as chasmolithic colonists and penetrate the fabric of porous, translucent rocks to provide the primary-producing basis of endolithic communities ranging from the hottest deserts to the cold Dry Valleys of Antarctica. They biodegrade these rocks to create soils which they enrich and inoculate. Their ability to survive at the limits of life on the surface of the Earth is now being studied as an analogue for past life on Mars ? the ultimate desert.” DNA repair genes.
Jessica – roadrunner https://en.wikipedia.org/wiki/Roadrunner . Able to fly, but prefers to run on the road – perhaps to conserve energy. During the cold desert night, the roadrunner lowers its body temperature slightly, going into a slight torpor to conserve energy. To warm itself during the day, the roadrunner exposes dark patches of skin on its back to the sun.[13]
Kostas – resurrection plant. Many plants, including https://en.wikipedia.org/wiki/Selaginella_lepidophylla . Springs back to life over 3 hours on exposure to water after dessication. Can lose roots and tumbleweed around. Flavinoids from these inhibit photosynth in spinach chloroplasts https://pubs.acs.org/doi/full/10.1021/jf8010432 .
Also, desert-adapted fungus https://en.wikipedia.org/wiki/Agaricus_deserticola – protective membrane when small, closed cap adaptations.
Kazeem – https://en.wikipedia.org/wiki/Xerophyte – including Agave https://en.wikipedia.org/wiki/Agave – back to https://www.sciencedirect.com/science/article/pii/S209012322300125X
Ramon (guest!) – Ecology and natural history of desert lizards by Eric R. Pianka
3. Hybrid vigour
Jessica – https://www.sciencedirect.com/science/article/pii/S1360138507001847 towards the molecular basis of heterosis. When an offspring of two different inbred parent lines shows increased performance relative to either parent. The dual of inbreeding depression. Classical example – maize. Potential mechanisms –
Dominance: Term from quantitative genetics explaining improved vigour of hybrid plants by the complementing action of superior dominant alleles from both parental inbred lines at multiple loci over the corresponding unfavourable alleles.
Epistasis: Term from quantitative genetics explaining the superior phenotypic expression of a trait in hybrids by interactions between non-allelic genes at two or more loci.
Overdominance: Term from quantitative genetics attributing superior hybrid traits compared to their parental inbred lines to allelic interactions at one or multiple loci.
Q: why so much focus on maize? Agricultural importance, regions of growth, history of discovery; ploidy?
Yusra – hybrid cotton production https://link.springer.com/article/10.1007/s00122-023-04334-w . Overall to produce hybrid lines, want to avoid self-pollination: must hence remove male fertility of one line. Methods – HEP – hand emasculation and pollination; CMS – mito features make males sterile; GMS – nuclear features makes males sterile.
casual [sic?] mitochondrial genes for G. harknessii CMS used commercially in hybrid seed production have been cloned (Wu et al. 2022, Ma et al. 2022, Mao et al. 2022, Xuan et al. 2022).
Lots of info about specific CMS lines for cotton.
Belén – dogs – outbreeding extends life expectancy (overcoming inbreeding depression). Other animal examples via Wikipedia: “Black Baldy” cattle, broiler poultry, dogs, humans (IQ?), birds
Kazeem – in Nigeria – some lines of goats with high growth rates, some with desirable height/weight characteristics – bring together. Soya bean – miRNA families conserved in CMS soybean. Other types of male sterility: genic, cytoplasmic-genic, chemically-induced, transgenic. https://www.frontiersin.org/articles/10.3389/fgene.2021.654146/full
Rajneesh – onion. https://dergipark.org.tr/tr/download/article-file/2933318 – odd journal? Hybrid vigour in onions useful for shelf life and storage, and disease resistance. Uniformity of size and colour also improved. Inbred lines hard to maintain because inbreeding depression is very pronounced.
Kostas – CMS in animals. Reported “for the first time” in snail https://pubmed.ncbi.nlm.nih.gov/35483362/ – but seemingly also in fly https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3182495/ ? Review of crops here https://academic.oup.com/hr/article/doi/10.1093/hr/uhab039/6510196
2. Concepts in evolution
Kostas – competitive exclusion and extinction, through the case study of steam locomotion in material culture https://royalsocietypublishing.org/doi/full/10.1098/rsos.221210
Set of requirements for conditions where extinction is due to competition
Tractive effort (TE) as a phenotype that tells us a tremendous amount about individuals; a database contains almost all individuals that ever existed
Questions about bio metaphor – when we start to die off, don’t we retain only the fittest individuals, rather than decreasing maximum?
Sergei Valverde and Ricard Solé – more on cultural and material evolution
Rajneesh – convergent evolution. Case study of gene expression in camera eye development in octopus and human https://genome.cshlp.org/content/14/8/1555.short
Convergent evolution is the parallel evolution of similar phenotypes in lineages for which the common ancestor didn’t have that phenotype. Pax6 is a key player. Lots of genes are expressed in octopus and human eye; more so than human eye vs human connective tissue. Not quite clear how this reports on the convergence – except to say that our common ancestor had a set of genes that could form an eye if appropriately expressed, so maybe it’s the regulatory pattern taking advantage of this palette (perhaps via Pax6) that is the thing that evolved in parallel.
Kazeem – genome compaction in Oikopleura. Micro vs macro evolutionary processes: macro involving divergent evolution (speciation). Larvacean – free-swimming tunicate (Daniel Chourrout at Sars thinks a lot about these). Macroevolution and larvae stage of O. dioica. Hermaphrodites. Feed on marine snow. Easy to experiment with, gene expression can be knocked down or out. Good model for genetic and embryological studies. Can we use Clustered-HyperHMM to identify features through evolutionary pathways and show how Oikopleura evolution tunicates late development?
Belén – A paper from Martin et al. on endosymbiotic interactions https://royalsocietypublishing.org/doi/10.1098/rstb.2014.0330. Lychines an excellent such example, where alga + fungus live together by giving each other a mutually beneficial support. An example of symbiogenesis, where a new species is risen after symbiotic associations. New phenotypic characteristics and the evolution modes of competition vs cooperation. Another example of successful symbiosis: mitochondria https://pubmed.ncbi.nlm.nih.gov/29112874/
Robert – obligatory and facultative endosymbiosis. Different modes and levels of mutualism/parasitism. Margulis theory about cilia being symbionts. Examples paramecium bursalia. Parakaryon myojinensis, a ‘parakaryote’ that shares features of both prokaryotes and eukaryotes. Other examples of endosymbionts where organelle genes move or remain.
Jessi – EvoDevo as a research field. Compare different development stages to infer and understand evolution patterns. Toolkit genes, ancient and conserved across eukaryotes. Study of the gene expression dynamics. A paper is the introduction to the topic. Gene duplication is not very frequent. There are facts that could show against duplication. Many animal toolkit proteins show independent evolution traces.
No Responses