1. Speaker’s choice: big cells, or sessile single cells
Belén — lichen. Fungi with algal symbionts – https://www.cambridge.org/core/journals/lichenologist/article/lichen-algae-the-photosynthetic-partners-in-lichen-symbioses/A4808B98986967A742EF0DCFF187A937 Typically single chloroplast with variable morphology; some become flattened against cell wall at cell divisions, some more central. https://en.wikipedia.org/wiki/Lichen . Interesting from our theory’s perspective – should we view these as fixed and exposed to an environment, or a parasite?
Rajneesh – diatoms https://en.wikipedia.org/wiki/Diatom . Non-motile. Major marine microalgae group. 20-50% of planetary oxygen generation. Central to urea cycle. Source of nutrition – with psychological benefits? High in beta carotene. Useful proxy for water quality, also pH, salinity etc https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1442-9993.1995.tb00522.x
Sara – some aurora work! And magnetohydrodynamic simulations via https://vimeo.com/266469665
Kostas – Tree of Life visualisation https://www.onezoom.org/ – then green seaweed https://en.wikipedia.org/wiki/Caulerpa_taxifolia . Large seaweed with 60cm fronds, stems etc – but all a single cell. Multiple nuclei. Reproduces sexually but an invasive version (bred for aquaria?) seems not to sexually reproduce https://www.tandfonline.com/doi/full/10.1080/09670260802343640 ? (Very bad invasive species). Population genetics incl chloroplast DNA here https://onlinelibrary.wiley.com/doi/full/10.1046/j.1365-294X.2002.01627.x
Honorary mentions:
Brefeldia maxima -> unicellular slime mold
Xenophyophorea -> ocean-bed, have an exoskeleton. Multiple nuclei. almost like tissues?
Gromia sphaerica -> agains with tests, makes trails (!)
Acetabularia -> green algae. 0.5-10 cm, rhizoid, multinucleate (in the rhizoid). too complex for a unicellular?
Robert – Valonia ventricosa https://en.wikipedia.org/wiki/Valonia_ventricosa , also multinucleated, big spherical vacuole. It grows by multiple divisions. Outer wall resembles normal cells. Many chloroplasts. Color depends on the ‘intensity’ of chloroplast numbers. Flexible structure -> easy to regrow
Iain – Thiomargarita namibiensis https://en.wikipedia.org/wiki/Thiomargarita_namibiensis unusually very big bacterium -> energy demanding. Solution: splits its structure into multiple compartments with many copies of its genome. Lane and Martin in https://www.nature.com/articles/nature09486 use this bacterium as an example for the development of multicellularity. Acquisition of mitochondria led to more “free time” for the organisms to do more complex stuff.
2. Lichens
Rajneesh – dog lichen https://en.wikipedia.org/wiki/Peltigera_canina . Note lichen roles in erosion, rock breakdown, etc. Also measure of air quality in different areas. 3-5cm across, white and grey-black. Readout of soil in a poor state. Sy,mbiont is cyanobacteria of genus Nostoc , like the Azolla water fern symbiont. https://en.wikipedia.org/wiki/Nostoc
Sophie – genetic differences between fungal part of lichen symbiont and ancestral fungus https://www.sciencedirect.com/science/article/pii/S136952661730211X . Can modern lichen fungus survive independently?
Kostas – mtDNA gene reduction in lichen fungi https://onlinelibrary.wiley.com/doi/10.1111/mec.14519 – lost atp9? But it’s transferred to the nucleus https://onlinelibrary.wiley.com/doi/full/10.1111/mec.16010 . Red Queen connection with lichen symbioses – https://onlinelibrary.wiley.com/doi/epdf/10.1002/ece3.8471 . Fungi found in lichens have lower substitution rates than free-living relatives. Effects on longevity too? Old man’s beard lichen https://en.wikipedia.org/wiki/Usnea – including some preserved in Baltic amber from 34 mya.
Robert – https://en.wikipedia.org/wiki/Trebouxia Trebouxia, common algal symbionts in lichens. Furthermore, Trebouxia can exist in its free-living form or in a lichen thallus as a photobiont partner with its fungi mycobiont.[7] The release or escape of alga zoospores from intact lichens is a source of free-living algae colonies or single free-living cells. Moreover, the same Trebouxia species can be associated with many mycobiont species or many Trebouxia strains can inhibit single lichen.[12][22][29][11][1] However, the maturation of the lichen could lead to the elimination of all Trebouxia strains except one.[5] Also, Trebouxia species are not selective towards their fungal symbionts while fungal species are very selective regarding their algae partners.[5] In areas where algae species are scarce, fungi are less selective and forms a symbiotic relationship with any Trebouxia species and later on switch to a more suitable algae species.[5] Some Trebouxia species are highly dependent on their fungal partners and cannot exist as independent organisms.[30][31] Fungi obtain nutrients through self parasitism or selectively harvesting old Trebouxia cells.[5] Trebouxia, on the other hand, provides 90% of its photosynthetic products to the mycoboint.[5] Pyrenoglobuli (lipid rich stores in the pyrenoid of Trebouxia) are used by the mycoboint for energy and water.[5]
First mtdna from symbiont https://www.nature.com/articles/s41598-019-44700-7
Honorable mentions by Robert – 1. Soredium, the asexual reproductive structure of some lichens and 2. https://www.science.org/doi/full/10.1126/science.353.6297.337 for the importance of a third symbiont (as second fungus) in most lichens, which is a type of yeast
Belén – Asterochloris— clay/family Cladonia https://pubmed.ncbi.nlm.nih.gov/16918555/
Green algal photobionts in the Asterochloris group of the genus Trebouxia, fungus called Cladonia Subtenuis. Resistance to abiotic factors and stresses, first to populate areas after fires etc. There are different levels of organization to estimate photosymbiont’s activity. The local environment is key to determine a successful symbiosis.
Cladonia arbuscula- https://www.nature.com/articles/ismej200963 Over 60% of the bacteria are α-proteobacteria, forming colonies. Lichens as self-contained complex ecosystems. Translocation of elements and nutrients is important for the lichen’s well-being. Bacteria are involved in nutrient cycling (nitrogen fixation) in the lichen thalli.
3. Mito structure in basal metazoa
Kostas – Filamentous green algae (Characea https://en.wikipedia.org/wiki/Chara_(alga) ) living in less oxygenated water https://link.springer.com/article/10.1007/s00709-004-0075-1 – mitochondria resemble those in primitive plants – fragmented, move on actin. Like leaves – squeezed in semi-regular gaps between tessellated chloroplasts. Often sessile, but move to where needed in the cell. In rapidly elongating cells, giant (vermiform) mitochondria observed. Disrupting cytoskeleton with drugs leads to vermiform encirclings of chloroplasts and other fusion behaviour. Another paper on pH dependence of mito positioning here https://link.springer.com/article/10.1007%2Fs00709-019-01392-0 – “mitochondria were moved away from the cortex when a previously acid region became alkaline and vice versa.“
Robert – Ulva https://onlinelibrary.wiley.com/doi/full/10.1111/pre.12132 . giant mitochondria change structure during parthenogenesis. Single cell case, reticulated yeast-like structure; 5-cell phase, less hyperfused but still lumpy, not really bacteria-like
3a. Continued – plus other taxa
Belén — Brown alga Mutiomo cylindricus
https://link.springer.com/article/10.1007/s00709-021-01679-1?utm_source=xmol&utm_medium=affiliate&utm_content=meta&utm_campaign=DDCN_1_GL01_metadata . General picture – brown algal mitochondria are tubular, but fragmented – maybe dozens in the cell. In female gametophytes, there is a highly fused state. Some more fragmented too. Role for even spreading?
Rajneesh – Porifera – some hint of individual “blob” mitochondria https://www.researchgate.net/publication/282566875_Phylum_Porifera/link/57a838d608ae455e85470bc4/download Fig 8.12
Kostas – “Detection and cytoplasmic localization of two different microtubule motor
proteins in basal epithelial cells of freshwater sponges” Protoplasma 1997. Radial plus tubular structure? Also https://link.springer.com/content/pdf/10.1007/BF00329447.pdf . Quite fixed in place. Quite fragmented, move along “spokey” cytoskeleton from nucleus to periphery and back. “Shuttling”. Possible oxygen transport – high at membrane, low at nucleus? Not colocalised with ER, when treated with some chemical compounds. Followup – https://link.springer.com/article/10.1007/BF01323610 from the same team of authors, but with more detailed and fancy images. Also some useful numbers about mito distribution and their motility in these freshwater sponges. Mitochondria move somewhat faster in retrograde direction (0.4-2.8 μm/s) than in anterograde (0.5-2.2 μm/s).


On the left, the cytoplasmic organization of Spongilla epithelial cells. On the right, fluorescence micrographs of mitochondria therein.
Robert – mtfr1 and dufd1 (also called mtfr2) linked to mitochondrial network structure https://pubmed.ncbi.nlm.nih.gov/20568109/ – how is this link manifest and what relationship evolutionarily does it illustrate?
4. Picoeukaryotes
Belén — diversity and variation in picoeukaryotes in Arctic fjords around Svalbard. https://link.springer.com/article/10.1007/s00300-011-1097-8 diversity varies inversely with biomass. Picoplankton > 50% of autotrophic production before and after seasonal blooms. Parasites play a large role!
Kostas – https://en.wikipedia.org/wiki/Ostreococcus_tauri – smallest free-living eukaryote. Single mitochondrion, single chloroplast. PT and MT genomes are compact and dense https://academic.oup.com/mbe/article/24/4/956/1011264?login=true – smallest PT genome of plants and algae.
Rajneesh – general picoeukaryotes – <3um size, very common, important in photosynthesis. Surfactants and sonification – essential parts of analysing picoeukaryotes. Phylogenetics ribosomal RNA analysis for diversity. Reports of surprising diversity in Antarctic https://www.nature.com/articles/35054537 and https://www.nature.com/articles/35054541 (rRNA) back to back in Nature. Flow cytometry and fluorescence https://www.int-res.com/articles/meps/122/m122p001.pdf and https://agupubs.onlinelibrary.wiley.com/doi/abs/10.1029/98jc01333 , HPLC and flow cyt https://www.int-res.com/articles/meps/68/m068p121.pdf
Sophie – history of Osreococcus https://www.nature.com/articles/370255a0.pdf
Iain – giant Mimivirus infecting tiny algae https://en.wikipedia.org/wiki/Chrysochromulina_ericina_virus and https://journals.asm.org/doi/full/10.1128/JVI.02446-20 from Bergen. Encodes, weirdly, energetic proteins – Krebs cycle and ETC complexes. Why? Maybe shift metabolic priorities to what the virus can use?
5. Mitochondrion-related organelles
Yusra – https://onlinelibrary.wiley.com/doi/10.1111/j.1550-7408.2009.00407.x hydrosomes and mitosomes, evolutionary origins of mitochondria.
Rajneesh – http://news.bbc.co.uk/2/hi/8609246.stm , animal without mito. Loriciferans – multicellular organisms with hydrogenosomes https://www.tandfonline.com/doi/pdf/10.1080/14772000.2014.943820?needAccess=true . Live in sediment in the absence of molecular oxygen. Up to 1mm long, recently discovered group.
Belen – importing macromolecules into mitochondria and MROs https://royalsocietypublishing.org/doi/full/10.1098/rstb.2009.0167 . Look at import of proteins and tRNAs. Hydrogenosomes lose DNA but retain ATP production, mitosomes lose ATP production too. Everything retains ability to produce Fe-S clusters
Kostas- https://en.wikipedia.org/wiki/Anaeramoeba an anaerobic amoeba ‘Anaeramoeba’.
Placement in the tree of life, they lack proper mito so they have mito related organelles, they have more powerful hydrogenosomes, some info about the protein import system of hydrogenosomes, they get more amount that hydrogenosomes in close species, also ATP production. Fig 2 differences between metamonas and anaeramoeba. Fig 3 related to Hypertraps – https://www.sciencedirect.com/science/article/pii/S0960982221013646
Sophie- intracellular parasite Mikrocytos with really reduced mitochondria, difficult to place in the ToL. Figure 1 attempt of phylogenetic tree. They produce energy. https://www.cell.com/fulltext/S0960-9822(13)00758-6
Iain- unicellular eukaryote (protist) without mito nor mito-like organelles. No capacity to produce ATP. Steals bacterial genes for social mobilisation in the cytosol (prob. by horizontal gene transport). No own machinery for doing metabolism. https://en.wikipedia.org/wiki/Monocercomonoides
Review on these topics https://www.nature.com/articles/nature04546
6. Examples of organelle DNA mutations that cause disease
Rajneesh – https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance AMR disseases, in growth/development, caused by mtDNA mutations. Mutations on ATP genes. ATP not able to be produced.
Antimicrobial Resistance: Implications and Costs (nih.gov)– Table 2, mortality rates of AMR.
Robert- not a human, mutations in cpDNA in Arabidopsis, plant species with a small genome. Arabidopsis thaliana – Wikipedia
Frontiers | Chloroplast DNA Copy Number Changes during Plant Development in Organelle DNA Polymerase Mutants | Plant Science (frontiersin.org)– plastid DNA copy number changes during plant growth and development. Comparison of 2 mutants. The mutant plant lines have T-DNA insertions in genes encoding the two organelle localized DNA polymerases (PolIA and PolIB controlling replication of oDNA). These polymerases are out of the organelle (is a nucleus-encoded gene), so not organelle DNA mutation.
Yusra-mtDNA and Mitochondrial Diseases | Learn Science at Scitable (nature.com) loss of vision in both eyes, associated with mtDNA homoplasmic mutations. Associations with aging and cancer.
Iain- map of human mito genome and the associated mito diseases, illustration: Morbidity map of the human mitochondrial genome. The map of the 16,569… | Download Scientific Diagram (researchgate.net) some diseases LHON, MELAS, MERRF. Particularly common: MELAS at 3243
Belén-https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2790545/#:~:text=Based%20on%20current%20research%2C%20we,deficient%20mitochondrial%20DNA%20repair%20mechanisms%2C mitochondrial abnormalities and mtDNA mutations are involved in the development and progression of multiple sclerosis (MS) in humans. mtDNA nt13708A variant increases the risk of MS. Genetic variants of complex I genes may influence the response of tissues to inflammation in the CNS. Discovered an association among common variants of the mitochondrial ND2 and ATP6 genes with both MS and systemic lupus erythematosus.Variations in mtDNA and nuclear-encoded mitochondrial protein genes may contribute to disease susceptibility in MS.
Abnormal mitochondrial dynamics (imbalance in mitochondrial fission and fusion) plays a key role in tissues affected by multiple sclerosis.
Robert – https://pubmed.ncbi.nlm.nih.gov/16026996/ a dog disease called spongiform leukoencephalomyelopathy. It affects their neural system, solely maternally inherited. It’s generated by a missense mutation in cytochrome b. Affected dogs showed elevated ratio of 3-OH butyrate to acetoacetic acid.
Rajneesh – https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance Mutations disturbing ATP production. Linked to antimicrobial resistance.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6929930/ a survey about the impact of antimicrobial resistance
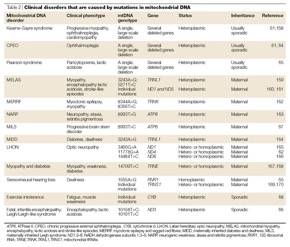
Kostas – first, in https://www.nature.com/articles/nrg1606/tables/2 there is a compact table with some known diseases caused by mtDNA mutations. Attached here for (quick) future reference. Note, this is from 2005, so it’s not complete and updated!
Next, in https://pubmed.ncbi.nlm.nih.gov/29940850/ a ptDNA mutation (in gene rps4) causes chinese cabbage to have dysfunctional photosynthesis and slow development. The mutation is homoplasmic and was shown to be maternally inherited. Tested also various combinations of crossings. Finally, this mutation possibly impairs the rRNA processing in that plant. Interesting point: Arabidopsis is a model organism for mitochondria , whereas tobacco is for plastid (because of the leaf size -> more plastids, as Iain pointed out). Worth following works by R. Bock on ptDNA, like the suggested by Iain -> https://academic.oup.com/plcell/article/28/4/911/6098150 with Table 1 therein of ptDNA mutants and their negative phenotypic effects. Also by Bock, the work in https://academic.oup.com/plcell/article/20/8/2221/6092598 shows a pt encoded gene is responsible for low-temperature tolerance in tobacco
6. Eukaryotes exhibiting antimicrobial resistance
Robert – https://journals.asm.org/doi/full/10.1128/AAC.01115-17
Leishmaniasis, serious disease and expensive/insufficent drugs. ABC Transporters are ubiquitous in eukaryotes and found on mito or plasma membranes. Leishmania have a huge set of those transporters. In this paper a new ABC transporter (ABCI3) only found in Leishmania major. Different life stages (promatigotes-early life stage before developing a flagela, and amastigotes-no flaggela) show different resistance levels. ABCI3 responsible for susceptibility to antimonial treatments and infectivity.
Honorable mention: Aspergillus fumigatus https://onlinelibrary.wiley.com/doi/full/10.1111/mmi.13167 They found that KO mgm1 disrupts normal fusion. Albeit reduced growth when messing with fission, they show resistance to a particular disease called aspergillosis and also azole resistance.
Yusra – http://eprints.gla.ac.uk/131520/1/131520.pdf A nice overview on drug resistance among eukaryotic microorganisms. Bacterial infections in eukaryotes and the high levels of resistance for the infecting bacteria. Development of resistance through different strategies, such as persister population. The persistence is a physiological state. Different mechanisms of persistence vary (bet hedging). A big challenge for clinical diagnosis and screening for drug resistance in eukaryotic pathogens. Multiple examples of diseases caused by such microorganisms and the status of drug resistance of them. Very interesting and compact figures showing (some) timelines and mechanisms of how drug resistance takes place (eg see Fig. 2).
Rajneesh – same paper with Yusra. A healthcare perspective on drug resistance. Focus on malaria, which is caused by apicomplexans. The most popular drug is chloroquine (also cheap).
Jo – https://aricjournal.biomedcentral.com/articles/10.1186/s13756-019-0559-6
A slightly different approach. This is a review of plants producing compounds that increase resistance of their pathogens.
Also https://journals.asm.org/doi/10.1128/AAC.02197-17 on staphylococcus.They showed tomatidine possesses potent antibacterial activity against Staphylococcus aureus by affecting the latter’s energy production
https://www.cambridge.org/core/journals/parasitology/article/tomatidine-promotes-the-inhibition-of-24alkylated-sterol-biosynthesis-and-mitochondrial-dysfunction-in-leishmania-amazonensis-promastigotes/B7698002E840439D19365C1DCCE2D790 again for tomatidine and how it affects the mitochondrial function in Leishmania amazonensis promastigotes.
Kostas – https://en.wikipedia.org/wiki/Somatic_fusion how we manipulate and enforce resistance to pathogens and pesticides by “crossing” somatic cells, to improve crops and production. For example https://link.springer.com/article/10.1007/BF00264975 https://pubmed.ncbi.nlm.nih.gov/24227155/ https://cris.bgu.ac.il/en/publications/intraspecific-transfer-of-herbicide-resistance-in-the-red-microal https://www.tandfonline.com/doi/abs/10.2216/i0031-8884-34-4-299.1
This also happens in nature through symbiotic relationships (and antagonism) and/or use of “exotic” organelles, eg https://link.springer.com/chapter/10.1007/978-3-030-60659-6_10. Also, in https://www.nature.com/articles/s41467-018-05651-1 where natural occurrence of extrachromosomal plasmids in red algae may be responsible for alga’s resistance.
7. Favourite prokaryote and eukaryote [after the summer break!]
Jessica – prokaryote Myxococcus xanthus: communication between cells, build multicellular biofilms with collective behaviour as a closed system when lack of food. Produce spores. Serve as food for the other bacteria in the proximity when they die. They are also predators of other microorganisms. Sequenced genome, longest known in bacteria. Some mutants ‘cheat’ without collaborating in the collective behaviour. https://academic.oup.com/femsre/article/36/1/149/534666 .
Iain – prokaryote: Klebsiella pneumoniae https://en.wikipedia.org/wiki/Klebsiella_pneumoniae and paper https://www.pnas.org/doi/10.1073/pnas.1501049112 . Normal flora and pathogenic strains that infect the lungs (areola? IGJ haha no, alveoli. An awkward slip from me – Google both if you don’t know the diff!). Presents multidrug resistance, potentially by plasmic transfer to other bacteria. Eukaryote: Dictyostelium discoideum https://en.wikipedia.org/wiki/Dictyostelium_discoideum exists as a single cell, but when environmental conditions get hard they aggregate and form multicellular structures. Big altruistic behaviour so that genes are passed to other members in the population. Honorable mentions: bumblebees, tent-making bats, tardigrades, horsetails, azolla, irish moss, and reclinomonas americana.
Robert – prokaryote: Thiomargarita https://en.wikipedia.org/wiki/Thiomargarita_magnifica, recently discovered, very unusual. Largest known bacteria, first thought of being a fungus because of its reproduction. They are highly polyploid (368000) with the genomes distributed all over the cytoplasm and they store a lot of water on the inside, these two strategies make them so big. Tick cytoplasm pushed against the cell’s wall. Genes involved in nitrogen production. Eukaryote: Monocercomonoides https://en.wikipedia.org/wiki/Monocercomonoides. Is a parasite. Some genus have flagella. Lack of mtDNA and lack of mitochondria-related organelles. Speculated that they had mitochondria in the past. Honorable mention for Parakaryon https://en.wikipedia.org/wiki/Parakaryon_myojinensis in-between prokaryotes and eukaryotes.
Rajneesh – prokaryote: Cyanobacteria. Gramm-negative photosynthetic bacteria. Blue or green pigments. They fix nitrogen from the atmosphere and also reduce the amount of carbon in the water. Block the sunlight from other organisms (which is harmful). They contaminate drinkable water. Paper about how to improve the water quality looking at several parameters: https://www.sciencedirect.com/science/article/pii/B9780128113301000016. Methods for controlling and detecting cyanobacteria: http://aqualab.com.au/wp-content/uploads/2017/10/Application-Note-Cyanobacteria-Detection-in-Water-using-in-vivo-Fluorometery-AUS.pdf. Eukaryote: cancer cells. Created when genes for regulating cells are damaged. Cancer cells do not stop growing and dividing, and they ignore signals from other cells. Ancestors of plastids.
Belén – prokaryote: Heimdallarchaeota, type of Asgard archaea https://en.wikipedia.org/wiki/Asgard_(archaea) . Presented in 2017. Closest relative of the ancestor of the nucleus of the eukaryotic cell. Figure 1 in https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6646929/ of Loki’s castle location and Aarhus where they found it. Fig 4 for two-domain classification of life. University of Bergen discovered Loki’s castle https://en.wikipedia.org/wiki/Loki%27s_Castle . Review of origin of eukaryotes: https://www.nature.com/articles/nature21031 . Eukaryote: rabbits https://en.wikipedia.org/wiki/Rabbit cannot cry nor vomit. Regulation of body temperature using their ears. ‘’Invasive species’’, check their origin in Europe.
Kostas – prokaryote: Halobacterium, is not a bacterium but an archaeon. Salty environments. Aerobic metabolism. Eukaryote: Naegleria gruberi, related to a deadly one that affects the brain. This one is harmless. Different life stages, from ameba to flagellated mobility structures. A lot of mitochondria and mt genes that encode for proteins, like humans (13).
8. Long-lived or unexpectedly motile organisms
Rajneesh – Turritopsis dohrnii: immortal (not ageing) jellyfish, mainly found in tropic/warm waters. Carnivorous, fed on zooplankton. Mouth used for both feeding and defecation. Has the ability to alternate between life stages. They reproduce both sexually and asexually.
https://www.journals.uchicago.edu/doi/pdfplus/10.2307/1543022 how they reverse their life cycle.
Well-studied for its immortality.
Belén – Nematostella: sessile, marine anemones. When they starve, they shrink (body, cells, etc). This helps them have really long life span. Relationship with a lot of symbionts. Cell proliferation when food around. Asexual and sexual reproduction? https://www.sciencedirect.com/science/article/pii/S1084952109001736?via%3Dihub for hydras, similar organisms to nematostella. Both have unlimited regeneration capabilities. Similarities with regeneration observed in plants.
Yusra – Planaria flatworms. Reproduce sexually and asexually. Quick regeneration of body parts.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5051648/ regeneration and inheritance in planaria
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3306686/ they maintain telomeres (one hallmark of ageing), in other organisms those degenerate as the organism ages, they are effectively immortal
https://pubmed.ncbi.nlm.nih.gov/19247944/ overview of research on planaria
https://www.nottingham.ac.uk/news/pressreleases/2012/february/immortal-worms-defy-ageing.aspx
Jessica – American lobster
https://www.sciencedirect.com/science/article/pii/S0531556509000849 again telomeres involved in the prolonged life span
Robert – Hydra: immortal Cnidarion found in freshwaters, high levels of regeneration, diploblastic, motile, they bud when food is abundant, DNA repair and telomeres involved in their immortality. They can detach themselves from the seabed and move. https://www.sciencedirect.com/science/article/pii/S096098221730338X and https://www.tandfonline.com/doi/full/10.1080/07924259.2014.927805 about how hydras die
Kostas — (Euplotes eurystomus), unicellular ciliate, uses 14 cirri (like legs) to walk in an unexpectedly sequential and orchestrated pattern
https://www.sciencedirect.com/science/article/pii/S0960982222011617
Orchestrated way of walking, they analyzed its gait and found 32 states. Gait followed a sequence transition among those states. Tubular, fiber-like bundles inside the cell is the mechanistic propeller of the movement. Looks like Strandbeest in https://www.youtube.com/watch?v=LewVEF2B_pM
9. Effective population sizes
Effective population size is a parameter of a genetic model (often Wright-Fisher). It is not directly linked to actual population size (number of organisms), and it is hard to estimate because in a sense it corresponds to a “spread” parameter. It affects behaviour like genetic drift rates, fixation times, etc.
Rajneesh – features linked to rate of gene evolution in eukaryotes https://academic.oup.com/gbe/article/14/2/evac028/6528851#334476154 . We find that the rate of adaptive evolution is significantly positively correlated to the rate of recombination, protein length and gene expression level, and negatively correlated to gene age. Only tangential link to Ne: Although, the effective population size in humans has increased recently, the effective population size is considerably reduced from that in the human–chimpanzee ancestor (Hobolth et al. 2007; Burgess and Yang 2008; Prado-Martinez et al. 2013; Schrago 2014)
Belen – https://academic.oup.com/gbe/article/4/5/658/533755?login=false (Table 2– Ne for several eukaryotes)
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1461962/pdf/11861575.pdf (Table 3 – Ne for several eukaryotes; Table 4 – outputs from simulation) – why are these lower? – probably subpedigrees
NB – Drosophila Ne is very different (10^5 vs 10^2) in the two papers
Ratios of real to effective population size for many multicellular eularyotes https://www.cambridge.org/core/services/aop-cambridge-core/content/view/59D48CF3CCD19ECE47163795F6EEA89E/S0016672300034455a.pdf/effective-population-sizeadult-population-size-ratios-in-wildlife-a-review.pdf
Often around 0.5? – probably subpopulations
Estimates of variance in Ne across genomic sites https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3241429/
Kostas – Estimates of mammals https://onlinelibrary.wiley.com/doi/10.1002/ece3.1249 – especially Appendix 9 https://onlinelibrary.wiley.com/action/downloadSupplement?doi=10.1002%2Fece3.1249&file=ece31249-sup-0009-AppendixS9.pdf (humans 10k)
PhD thesis – https://core.ac.uk/download/pdf/154430709.pdf p5 with lots of estimates Table 1.1
Leads us to Lynch 2016 – https://www.nature.com/articles/nrg.2016.104#MOESM6
See Supp Table – generally humans 20k, other multicell euks 100s k, unicell euks 1-10m

10. Gene transfer (organelles, eukaryotes, prokaryotes)
Belén – Nuclear pores: holes on cell’s membrane. https://www.ncbi.nlm.nih.gov/books/NBK9927/ About the nuclear pore complexes and how they enable gene transferability. See also fig 8.5
Also https://www.sciencedirect.com/science/article/pii/S0168952501023125?via%3Dihub about mito genes going into the nucleus. Directly or via intermediate stages (complementary DNA). In fig 1 the most common mechanisms of transfer of mito DNA to the cell’s nucleus. Direct transfer of mtDNA is more common in higher plants.
Yusra – https://www.tandfonline.com/doi/full/10.3109/1040841X.2013.804031 Horizontal gene transfer from E. coli to humans and description of the three most common ways it takes place (Conjugation, transformation, transduction). Not always with pathogenic effects. A video https://www.youtube.com/watch?v=08Q-MVeNeTU&ab_channel=NeuralAcademy explaining the three ways HGT takes place.
https://www.frontiersin.org/articles/10.3389/fpls.2017.02015/full#:~:text=Most%20examples%20to%20date%20include,into%20their%20host%20plant%20genomes HGT in plants. Agrobacterium, a prokaryote involved in massive HGT to plants. We use agrobacterium to edit plants’ DNA in labs.
Jessica – Viruses contribute to HGT. Retrovirus introduce their DNA to cell’s DNA. iain’s remark: we own the formation of placenta to retroviruses
Rajneesh – https://www.cell.com/action/showPdf?pii=S0168-9525%2821%2900051-2 HGT contributes to a bigger genetic pool. HGT in fish that helped them battle freezing water by lowering their blood freezing temperature. Description of how that event took place. First documentation of HGT between vertebrates. The postulated physical mechanism is by DNA attaching to free-floating sperm. https://www.the-scientist.com/slideshows/slideshow-examples-of-eukaryotic-horizontal-gene-transfer-70130 a slideshow with examples of HGT in eukaryotes.
Iain – Transfer of MSH1. MSH1 has made it to mitos of some species. Beneficial transfer, thus retained and selected for. Potential source of the gene is an alga, the transfer vector was probably a virus https://pubmed.ncbi.nlm.nih.gov/21801381/
Also, use of https://en.wikipedia.org/wiki/Bacillus_thuringiensis for genetic engineering to infuse plants with insect tolerance
Kostas – An very brief overview of why HGT is more common in prokaryotes than eukaryotes. Use of viruses and viral vectors to force gene transfer, also used in gene therapy. Targeted at germline rather than somatic cells. Iain’s comment: grafting is also a means for HGT, link to Bock’s paper about the ‘donation’ of plastid https://www.science.org/doi/10.1126/sciadv.abd8215 via grafting. Grafting is nice mechanism to enrich HGT and even organelle exchange, as Iain’s example notes.
11. Network structures in biology
Kostas – biological network growth in complex environments https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1008003 (focussed on cells, but multiscale – general generating process under external signals) – biased correlated random walk
Fibroblast reticular cell network – https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1002515 – relevant for lymph nodes, where cells form small world networks whose smallworldness is robust to edge removal. Assessment of topological robustness from real-world data. Key control parameters – are these inferred or guessed?
ER and microtubules https://www.pnas.org/doi/10.1073/pnas.2104309119 . Analysis of ER network using “CellProfiler” – showing a (causal?) link to microtubule distribution. Modelling of ER.
Iain – ER structure and optimality. Construct MST for moving compartments in the cell – get something that statistically looks like ER structure. Kostas – also https://en.wikipedia.org/wiki/Relative_neighborhood_graph and Gabriel graph https://en.wikipedia.org/wiki/Gabriel_graph – various subset relationships; https://en.wikipedia.org/wiki/Urquhart_graph
Rajneesh – metabolic networks https://en.wikipedia.org/wiki/Metabolic_network . Catabolism – obtaining energy; anabolism – production of new cell components. Focus on glycolysis – producing ATP. 10 steps – preparatory and payoff reactions. Metabolic networks and their respective fluxes can distinguish disease states, tissues, species, etc
Robert – tortoise-shell bamboo https://en.wikipedia.org/wiki/Phyllostachys_edulis . Root systems characterised with ground-penetrating radar https://www.mdpi.com/2072-4292/13/14/2816 . Root systems as an example of one-way transport systems – other plants also have great data on this (eg transparent soil)
Belen – dynamic ER behaviour https://www.cell.com/biophysj/fulltext/S0006-3495(14)00675-4 (Imogen Sparkes)
Dynamic elements remodel around static ones. “Euclidean Steiner tree” picture. Steiner points are degree 3 and angle 120deg, flexible points aren’t – just wiggly ends? Langevin dynamics for wiggly ends – what is the drift term?
ER contacts with other organelles https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3683900/ – especially ER-mito contact sites
And Mark Fricker’s paper on plant ER dynamics https://www.nature.com/articles/s41467-019-08893-9 quantitative structure, topology, etc
No Responses