1. Freestyle
Belén — flatworms [also note Fasciola hepatica with reduced mitochondria]
Kostas — polychaete worms Nereis pelagica and the bivalve Arctica islandica [colder environment -> lower oxygen uptake, effects on lifespan?] [these mitos take up more oxygen when PO2 is higher; cow heart mitos don’t] https://jeb.biologists.org/content/203/21/3355.short
Robert — Silene conica (sand corn catchfly) and Silene noctiflora [massive, multichromosomal mtDNAs that gain and lose whole chromosomes]
Iain — Tachyglossidae (echidnas)[odd physiological mating strategies, surprisingly low metabolism, quite good at environmental stress] [also https://en.wikipedia.org/wiki/Carcinisation came up — convergent crustacean evolution to crab-like forms]
Jo — Xestospongia muta (giant barrel sponge) https://en.wikipedia.org/wiki/Giant_barrel_sponge [very long lived]
Arunas — Apis mellifera capensis (Cape honey bee) https://en.wikipedia.org/wiki/Cape_honey_bee [uses thelytoky — females are produced from unfertilised eggs — controlled by a single gene. Also employs social parasitism, where the parthenogenic offspring of Cape bees take over another bee colony]
2. Variegation
Jo — tea (Camellia sinensis) 0.01% plants in plantation, variegated by branch and by site in leaf, omics show links to expression of chlorophyll, ribosome, and PSII related genes https://pubmed.ncbi.nlm.nih.gov/27633059/
Belén — Cyclamen purpurascens variegated leaves either by border or vein https://en.wikipedia.org/wiki/Cyclamen_purpurascens
Kostas — common fig (Ficus carica) https://en.wikipedia.org/wiki/Common_fig variegation by leaf border / half with transcriptome profile more protein synthesis, less degradation https://www.mdpi.com/1422-0067/20/6/1338/htm — suggested variegation can be beneficial? Predator avoidance? variegation also because of viruses, eg leaf mottling in Daphne odora.
Robert — Pilea cadierei (aluminium plant) — striking stripy leaves, but not clear if this is genetic variegation or just air pockets? “Genetic” vs “chimeric” variegation. Zea mays stripe mutants caused by mtDNA mutation https://www.ncbi.nlm.nih.gov/pmc/articles/PMC386717/pdf/pnas00323-0243.pdf “sortingoutofnormalandmutantmitochondriainaffectedplantscanresultinsometotallynormalprogeny,butgermcelllineagescontainingonlymutantmitochondriawouldgivenoviableprogeny.”
Iain — Ilex (holly) especially at Kenilworth castle where whole branches are yellow and the rest are variegated by leaf border
3. Organisms with msh1
Jo — Emiliania huxleyi — coccolith. Msh1 identified from a metagenomic study!? https://www.pnas.org/content/117/28/16448 other interesting clades here too. Worth BLASTing grene plant msh1 against huxleyi reference genome? https://www.ncbi.nlm.nih.gov/genome/?term=txid2903[orgn]
Kostas — corals and specifically bubblegum coral https://en.wikipedia.org/wiki/Paragorgia_arborea via https://www.sciencedirect.com/science/article/pii/S1055790309005132 . Deeper corals have larger growth / body plans? Link to gene retention? (sea pens also live deep)
4. Phenotypic plasticity
Jo — heterophylly (leaf form plasticity) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5632738/ Terrestrial and aquatic species — Potamogeton nodosus, Ludwigia arcuata, and Rorippa aquatica. What scale of environmental cues shape these effects? Single leaf / whole organism / both?
Belén — Sylvilagus nuttallii (mountain cottontail rabbit) and Lepus americanus (snowshoe hare) — https://royalsocietypublishing.org/doi/pdf/10.1098/rspb.2014.0029 seasonal changes in coat colour, but limited plasticity (so can’t keep up with environmental change). Could model this — delayed, oscillatory predation dynamics?
Kostas — Paris polyphylla (Asian flowering plant — no common name?) many varieties, different leaf shapes, diploid vs tetraploid, etc https://www.sciencedirect.com/science/article/abs/pii/S1874391919300466
Iain — Daphnia (water fleas) — grow spikes and grow faster in face of chemical cues in the water https://natureecoevocommunity.nature.com/posts/28994-can-we-detect-local-adaptation-of-phenotypic-plasticity
5. Carnivorous plants
Robert — Drosera spp. (Sundew) —https://en.wikipedia.org/wiki/Drosera_capensis Leaves grow with cell size tension, so curl on touch. Capture prey with sticky trichomes. Self-pollinating.
https://www.mdpi.com/1422-0067/20/17/4107/html Plastome contains 97 genes, has all photosynthetic genes but “The most notable loss is the lack of all 11 genes coding for the NAD(P)H dehydrogenase complex (ndhA–K). Also missing are genes ycf1 and ycf2, thought to be essential in plant plastids.”
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5026580/ Novel proteases found in Drosera species.
Kostas — Utricularia (bladderworts) — https://en.wikipedia.org/wiki/Utricularia — vacuum mechanism to capture prey triggered by entrance of bug (has false alarm switch too). Millisecond timeframe. Eat algae too https://link.springer.com/article/10.1007/s11258-008-9420-3
https://www.sciencedirect.com/science/article/abs/pii/S030437700400138X Cost and benefit analysis of carnivory/nutrient (N and C) gain in bladderworts. When environment Lacks N, plant invests (grow bigger and more bladders) to capture more prey. When environment lacks C, not as much investment. N is most limiting.
— general notes — Most carnivorous plants lack roots, relying on rhizoids, don’t have proper structures. (most likely as they live in nutrient sparse environments). They still use photosynthesis, perhaps even more than other plants- they can reduce their energy consumption (hibernation-like state) if they don’t get enough light. You need to develop a particular strategy and make use of the prey to be a truly carnivorous plant. Independent appearance of carnivorous plants has happened 6 times (convergent evolution). Fossil record difficulty as they are fleshy, not woody.
Belén — Darlingtonia californica (pitcher plant) — https://en.wikipedia.org/wiki/Darlingtonia_californica attract prey with nectar, tubular leaves trap prey. This shape reduces surface area with chlorophyll, so rely on bright sunlight for what is left. Sole member of Darlingtonia, grow in nutrient poor freshwater environments. Dormant in winter. Highly modified root systems, can help sustain plant after fire. Big gap in temperature between what root and stem can tolerate, root much lower around 10 degrees C. https://web.archive.org/web/20190531212802/http://www.esalq.usp.br/lepse/imgs/conteudo_thumb/The-roots-of-carnivorous-plants.pdf
Jo –– Genlisea spp. — https://en.wikipedia.org/wiki/Genlisea Traps protozoa, invertebrates, algae, with its corkscrew-like structure with 400µm x 180µm openings, video of an annelid going in: https://www.youtube.com/watch?v=k4ETYSCRO7o
Also this paper on plastid genome https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0190321
6. Possible archaeal precursors to eukaryotes
Jo — Isolation and culture of an Asgardian https://www.nature.com/articles/s41586-019-1916-6 — hypothesising about eukaryogenesis (Fig 5). Parallel question — how did nuclear envelope evolve? https://link.springer.com/article/10.1134/S1990747816030156
Kostas — archael biofilms https://www.nature.com/articles/s41579-018-0058-4#:~:text=Biofilms%20containing%20archaea%20have%20also,within%20these%20biofilm%20communities28. Looks like similar principles may underlie bacterial and archael biofilms. Collectivist parallels between bacteria and mitochondria https://www.frontiersin.org/articles/10.3389/fphys.2019.00340/full
[ parallel discussion — large-scale “identification” of many many species from the Costa Rican jungle https://zookeys.pensoft.net/article/55600/element/4/444// — and associated controversy https://onlinelibrary.wiley.com/doi/10.1111/syen.12444 ]
Belén — alpha-proteobacteria like Rickettsiales https://en.wikipedia.org/wiki/Rickettsiales look like what the proto-mitochondrion may have looked like. Phylogenies for alpha-proteobacteria https://jb.asm.org/content/189/13/4578 . Also Nanoarchaeum equitans — smallest genome ever (491kb) and only known archaeal parasite https://www.pnas.org/content/100/22/12984
7. Unusual sensing mechanisms (in sessile organisms)
Jo — brown algae and mechanosensing. Plant mechanosensing https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-017-0403-5 (Olivier Hamant); suggested to guide morphogenesis in brown algae too, e.g. in response to wave stress https://pubmed.ncbi.nlm.nih.gov/22513108/ . Can increase fastening, adjust width of “leaf” blades, … . What sort of timescales are we talking? Perhaps morphogenesis is quite slow compared to sensing, requiring integration of the signal(s)? https://www.frontiersin.org/articles/10.3389/fpls.2014.00471/full ; also tip growth https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.2005258
Belén — gravitropism in plants, and stochastic influences https://www.frontiersin.org/articles/10.3389/fpls.2014.00674/full . Thermal and mechanical noise, which sensing must overcome. Chara as an example alga https://en.wikipedia.org/wiki/Chara_(alga) . Poplar gravitropism https://pubmed.ncbi.nlm.nih.gov/19453506/
Kostas — Sicyos angulatus (star cucumber) — “thigmomorphogenesis” — morphogenesis in response to touch — can sense mass of 0.5g (humans sense 2g). Also — how roots recognise self vs non-self https://www.pnas.org/content/101/11/3863.short . This may at least in part be via secreted chemicals https://www.tandfonline.com/doi/full/10.4161/cib.3.1.10118
Iain — seeds sense constant vs varying temperature, integrating a signal via physical separation of responses https://www.pnas.org/content/114/25/6629
8. Alien life / life in space
Jo — https://www.frontiersin.org/articles/10.3389/fmicb.2019.00780/full — extremophiles and the planetary context of life
Iain — https://www.popularmechanics.com/space/deep-space/g1592/we-asked-7-experts-what-would-aliens-actually-look-like/ — whimsical pop article about what aliens might look like — to critique! Clear anthropocentrism and some interesting ideas
Belén — tardigrades! https://en.wikipedia.org/wiki/Tardigrade — lower < 0.01% metabolism, survive very diverse extreme conditions. Have survived outer space! BIOPAN — organisms exposed to outer space https://en.wikipedia.org/wiki/List_of_microorganisms_tested_in_outer_space . Also a summary paper https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2832349/ ; which organisms survived space features; which survived panspermia-like impacts
Kostas — Halicephalobus mephisto https://en.wikipedia.org/wiki/Halicephalobus_mephisto nematodes found deep (2km) under the Earth’s crust, heat-adapted genome https://www.nature.com/articles/s41467-019-13245-8 including expanded Hsp arsenal . Also https://en.wikipedia.org/wiki/Chroococcidiopsis as a potential terraforming organism for Mars (extremophile phototroph); also https://www.asmscience.org/content/journal/microbe/10.1128/microbe.1.120.1
Sophie — podcast https://www.bigbiology.org/rss-feed/2018/9/21/ve29d5x8hcdq38kfphinj2hajpjrn3 — Sara Walker astrobiologist on “what is life”; https://www.cambridge.org/core/journals/international-journal-of-astrobiology/article/abs/on-the-probability-of-habitable-planets/B3B5368C97F1E15B0AA162E7CE47E999 probability of habitable planets; and https://www.cambridge.org/core/journals/international-journal-of-astrobiology/article/abs/organic-host-analogues-and-the-search-for-life-on-mars/183F1E399ADB23DFE855274264A32650 on life on Mars — are there Earth habitats that could “model” the Martian environment?
9. Unusual symbioses
Sophie — ants farming aphids. Aphids also harbour https://en.wikipedia.org/wiki/Buchnera_(bacterium) in obligate symbiosis. Also a bacterium https://en.wikipedia.org/wiki/Hamiltonella_defensa that provides resistance to wasp parasitoids, but only when it is itself infected by a phage!
Jo — Symbiodinium https://en.wikipedia.org/wiki/Symbiodinium — dinoflagellate symbionts of corals, sea anemones, jellyfish, flatworms, ciliates etc. Common in coral reefs. Exchange photosynthetic products for inorganic molecules. Can be endosymbiosed. Taxonomy recently reorganised https://www.cell.com/current-biology/fulltext/S0960-9822(18)30907-2?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS0960982218309072%3Fshowall%3Dtrue Rich ecology — different niches filled by different variations, many expelled into environment during business as usual by e.g. cnidarians. Interestingly, they have a single reticulated chloroplast that seems to span the periphery of the cell.
Also, seem to confer some robustness to climate change — population shifts under increased temperature allow host corals to survive https://royalsocietypublishing.org/doi/10.1098/rspb.2006.3567
Belén — cyanobacterium and unicellular alga https://science.sciencemag.org/content/337/6101/1546.abstract — that paper calls the bacterium UCYN-A and the alga is a prymnesiophyte. Exchanges nitrogen for carbon; acts as a nitrogen fixer, lacks PSB and TCA. From 2012 — similarities with the 2021 discovery of an endosymbiont allowing its ciliate host to “breathe” nitrate https://www.nature.com/articles/s41586-021-03297-6
Kostas — creosote gall midge https://en.wikipedia.org/wiki/Creosote_gall_midge . Quite parasitic wrt host plant, more mutualistic with a fungus. Induces a gall, inside which a fungus grows, which is eaten by the offspring. Monograph on its biology https://onlinelibrary.wiley.com/doi/full/10.1111/j.1479-8298.2012.00539.x . Some evidence that they prefer host species with different ploidies https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6749999/pdf/nihms-997455.pdf
Iain — Candidatus Azoamicus ciliaticola nitrate-metabolising endosymbiont found in a ciliate at the anoxic bottom of a nitrate-rich lake https://www.nature.com/articles/s41586-021-03297-6 — has ETC, precursor, ATP synthase, etc. Claim that host has hydrogenosomes.
10. Seaweeds
Jo — Ectocarpus brown alga, small, “frilly” (filamentous), anchored to sea bed. https://en.wikipedia.org/wiki/Ectocarpus . Useful review of seaweed lifestyles https://www.degruyter.com/document/doi/10.1515/bot-2016-0091/html — Ectocarpus among the most complex. Spores -> gametophytes -> (identical) gametes. Gametes can choose asexual or sexual routes. Parthenogenesis can go down haploid or diploid routes to produce next spores. Link back to Keeling’s plastid review https://royalsocietypublishing.org/doi/10.1098/rstb.2009.0103 — Ectocarpus (stramenopile) have undergone secondary endosymbiosis and thus have red algal-derived plastids. Inheritance of organelles — https://www.nature.com/articles/s41598-020-58817-7 — biparental inheritance of plastids, not mitochondria (maternal, with some paternal instances). Isogamy — plastids retained from both identical gametes, paternal mitos degraded.
Robert — Ectocarpus siliculosis, develop singly from terminal cells from lateral branches, dividing transversely. “plurilocular gametangia” as part of the development and inheritance anatomy. Different lifestyles as response to difficulty of finding a partner for sexual reproduction in ocean-released reproduction. Add: Case study suggest protist isogametes carry distinct genes related to sex, suggesting functional anisogamy precedes morphological differentiation into egg/sperm
Sophie — Zostera marina, eelgrass https://en.wikipedia.org/wiki/Zostera_marina ; drift and selection in clonal colonies following “belowground” proliferation https://www.nature.com/articles/s41559-020-1196-4 . Segregation of mutations into different branches, allowing “new organisms” to form from the clonal colony. Nuclear analogue of lots of our organelle work!
Kostas — Sargassum https://en.wikipedia.org/wiki/Sargassum , brown algae, oogamous, in Fucales (named for Sargasson sea). Some have lost their holdfasts and float for their entire lifespan. Paternally inherited centrioles https://link.springer.com/article/10.1007/s10265-005-0244-0 — microtubule association, influences degradation of organelles. Odd dance of nuclei in oogonium https://www.tandfonline.com/doi/abs/10.2216/i0031-8884-40-5-411.1 — retained longer than in other sisters, and plants etc. Fucales highly diverse group — many different reproductive strategies.
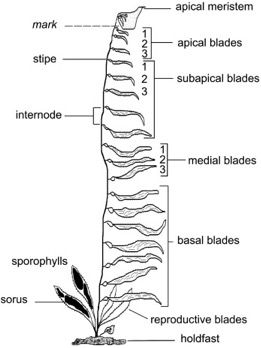
Iain — checklist — https://en.wikipedia.org/wiki/Fucus_vesiculosus (bladderwrack, brown) https://en.wikipedia.org/wiki/Chondrus_crispus (Irish moss, red), but also kelp and particularly https://en.wikipedia.org/wiki/Macrocystis . Kelp can be huge (60m) and fast growing (0.5m / day). Looks like reproductive blades are close to the bottom and generated by different meristem from the shoot apical meristem at the top, building solar panels (fig next page from http://www.bedim.cl/publications/cerdaetal_jembe_2009.pdf ) . So we have different reproductive strategies, but different developmental-reproductive strategies too: some algae generate fertile blades through the shoot apical meristem, some through a different meristem, and sea grass generates new clones via the “roots” (see Sophie).
11. Gold algae
Robert — Rhizochromulina . In SAR. One species (marina). Coloured amoeboids with single flagellum (although zoopores have two flagella). Their class — Dictyochophyceae — https://onlinelibrary.wiley.com/doi/full/10.1111/jpy.12904?casa_token=jDLy3fKmCgsAAAAA%3A9g1vsjvRmLLPqdrkb1DnPO_6Ix7CcTDwXW2pvXxJhYNm7aqJLr_tln4jseNebU4aVTF3DDfnpiA8Rwk — have “unusual” variability in their organisation. Repeat regions differ through the class. Quite a lot of gene loss — not immediately phylogenetically coupled. UiO thesis https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&ved=2ahUKEwjowbrd8fnvAhV7h_0HHd2iDHs4ChAWMAR6BAgFEAM&url=https%3A%2F%2Fwww.duo.uio.no%2Fbitstream%2Fhandle%2F10852%2F46108%2FAnders-Wold—Master-thesis.pdf%3Fsequence%3D1%26isAllowed%3Dy&usg=AOvVaw3rQjY6W0hZR92MLSdS32Tm
Cultured algae from Saudi Arabia and made some potentially new species discoveries? Differences in plastid count in different lifestyle stages? Some have stigmata.
Jo — Synurids https://en.wikipedia.org/wiki/Synurid — scale-like structures built on outside of chloroplasts, then wrap the cell? Freshwater organisms which form colonies? https://www.tandfonline.com/doi/pdf/10.1080/00071619000650111?needAccess=true more photos. Also — light-sensitive flagellum https://www.algaebase.org/search/genus/detail/?genus_id=43807 ? Do they move towards or away from light? “No stigma, but the smooth flagellum has a flagellar swelling acting as a photoreceptor, and colonies are strongly phototactic.”
Kostas — also synurids — colonies are heterothallic (sexes that reside in different organisms). Upon splitting, each daughter inherits some scales from the parent (binomial / other?). Also Chilomonas https://en.wikipedia.org/wiki/Chilomonas — golden-brown heterotroph with leucoplasts. They also contain nucleomorphs. Not known to sexually reproduce. Perhaps only non-photosynthetic member? Most have one or two chloroplasts.
Belen–Chrysochromulina. Not a typical golden algae- it’s macroscopic. Multicellular, but cells not physically connected (can slide away/be released). Lives in cold environments.
https://link.springer.com/article/10.1007/s10811-016-1047-5 Figure 4-7 shows morphology. Disturbance can release zoospores.
https://www.tandfonline.com/doi/full/10.1080/09670262.2011.598950 H.foetidus has an evolutionary origin in golden algae, they struggle to pinpoint H.foetidus amongst the golden algae, so is a clade of its own, its closest relatives are just samples from for eg baltic sea (unclassified)
Sophie– Dinobryon. Single celled, two differently sized plastids and two flagella, have an eye spot. Surrounded by a shell-like structure, https://link.springer.com/content/pdf/10.1007/BF01279321.pdf Can see daughter cell rotating in shell after division. Morphological isogamy, so has gender but look morphologically the same.
12. Red algae
Kostas — Cyanidioschyzon merolae — unicellular haploid adapted to hot sulphur springs https://en.wikipedia.org/wiki/Cyanidioschyzon_merolae , also https://academic.oup.com/pcp/advance-article/doi/10.1093/pcp/pcab052/6219305?login=true . Actually appear blue/green. >5k nuclear genes (high relative to other red algae). High GC content, 208 protein-coding ptDNA genes (150kb), 34 mtDNA genes (32kb). Single mitochondria and single plastid per cell. ptDNA smaller than other red algae, but codes more genes (barely any introns). Clever synchronisation for accurate choreography at cell divisions. FtsZ controls mito fission — also perhaps plastid, and whole cell? Genome reported https://www.nature.com/articles/nature02398 , ptDNA https://academic.oup.com/dnaresearch/article/10/2/67/371320 . Division machinery review https://www.nature.com/articles/s41586-021-03214-x.pdf — Iain adds that a recent paper highlights the role for membrane tension in mito division https://www.cell.com/cell-reports/fulltext/S2211-1247(21)00261-8
Sophie — “Coralline” algae https://en.wikipedia.org/wiki/Coralline_algae multicellular, reddish, purple, pink, other colours of algae populating coral reefs and growing on rocks. Strategies to avoid fouling. One species: Pneophyllum cetinaensis sp. Nov. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4726424/pdf/srep19642.pdf totally adapted to freshwater. Nice example of where retreating coast line led to a more freshwater environment developing over time, to which this sp adapted. Aside: marine -> fresh transition is rare (all fresh sponges descend from one marine species)
Iain — the common ancestor of most Rhodophyta , e.g. here https://deenr.rutgers.edu/Huan_Qiu_red_algae.html and from https://onlinelibrary.wiley.com/doi/abs/10.1111/jpy.12294 . Most red algae are descended from a single lineage via a tight bottleneck, and that lineage had lost 25% of its core genes and lots of its evolutionary plasticity. (Why?) Exceptions are in the Cyanidiophyceae like Kostas’ above. Maybe this is why green phototrophs dominate. NB — although most Rhodophyta have inherited this limited nuclear gene complement, ptDNA counts are still very high! Perhaps these are totally essential? Perhaps this is because red algae typically live in very dynamic (light) environments, e.g. beaches, tidal zone. What about Kostas’ extremophiles?
13. Environmental selection on organelle genetics
Robert — Tigriopus californicus. https://onlinelibrary.wiley.com/doi/full/10.1111/evo.12254 Survivorship measured among three strains plus hybrids after heat stress experiment. Apparently mtDNA evolution in these things is especially fast. Hybrids seem to show some hybrid vigour wrt surviving heat stress, and amount of vigour varies according to (cytb) mitotype.
https://onlinelibrary.wiley.com/doi/full/10.1111/j.1558-5646.2010.01135.x metabolic rate depends on mtDNA features and nuclear-mito crosstalk in seed beetles (incl Damian Dowling)
Jo — Haberlea rhodopensis (desiccating plant) https://www.frontiersin.org/articles/10.3389/fpls.2017.00204/full cpDNA analysis . Comparative analysis across relatives suggest that rps12 and rps23 seem to show some positive selection. Possible facilitators of quick changes for desiccation response.
Kostas — Rubisco across photoautotrophs https://bmcecolevol.biomedcentral.com/articles/10.1186/1471-2148-7-73 . Motivated by the idea that systematicists use rbcL without thinking much about whether it may be under different pressures etc. Evidence for positive selection in land plants, but less so in aquatic plants and algae and cyanobacteria (just low n? Written in 2007?). A few bases are responsible for most of the selective signal. More general review on organelle influences on plant adaptation here https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3172827/
Not sequence, but stoichiometry: more mtDNA in Chinese humans further north https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0079536 (is this just haplotype makeup?) — link to thermogenesis?
Also voles! https://www.nature.com/articles/hdy201428
Belén — Atlantic salmon. https://gsejournal.biomedcentral.com/articles/10.1186/s12711-015-0138-0 Some mtDNA features restricted to different latitudes. Role for temperature?
Iain — (mostly) human ideas and debate, including
Raquel Moreno-Loshuertos, Rebeca Acı́n-Pérez, Patricio Fernández-Silva, Nieves Movilla, Acisclo Pérez-Martos, Santiago Rodriguez de Cordoba, M Esther Gallardo, and José Antonio Enrı́quez. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nature genetics, 38(11):1261, 2006.
David C Samuels, Andrew D Carothers, Robin Horton, and Patrick F Chinnery. The power to detect disease associations with mitochondrial DNA haplogroups. The American Journal of Human Genetics, 78(4):713–720, 2006.
Pierre U Blier, France Dufresne, and Ronald S Burton. Natural selection and the evolution of mtDNA-encoded peptides: evidence for intergenomic co-adaptation. TRENDS in Genetics, 17(7):400–406, 2001.
Eduardo Ruiz-Pesini, Dan Mishmar, Martin Brandon, Vincent Procaccio, and Douglas C Wallace. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science, 303(5655):223–226, 2004.
Yongjun Luo, Xiaohong Yang, and Yuqi Gao. Mitochondrial DNA response to high altitude: a new perspective on high-altitude adaptation. Mitochondrial DNA, 24(4):313–319, 2013.
Dan Mishmar, Eduardo Ruiz-Pesini, Pawel Golik, Vincent Macaulay, Andrew G Clark, Seyed Hosseini, Martin Brandon, Kirk Easley, Estella Chen, Michael D Brown, et al. Natural selection shaped regional mtDNA variation in humans. Proceedings of the National Academy of Sciences, 100(1):171–176, 2003.
14. Mitochondria moving because of intrinsic properties
Jo — https://www.pnas.org/content/108/37/15456 — Ca2+ regulates mitochondrial motion in mouse hippocampal neurons. More Ca2+ — lower average speed (at the individual mitochondrial level). Ca2+ recruits Miro1 and “derails” mitochondria from the cytoskeleton. Added: In this study, Ca2+ level of mitochondria had no effect on directionality of movement, just mobility and speed.
Kostas — review paper from 2007 on mitochondrial motion https://www.sciencedirect.com/science/article/abs/pii/S096289240700164X . Does the mechanism “chosen” for mitochondrial motion depend on the distance they need to cover? e.g. yeast (short) vs axons (long)? Drosophila paper https://journals.biologists.com/jcs/article/119/4/659/29292/Regulation-of-mitochondria-distribution-by-RhoA suggests role for RhoA, which is a GTPase.
Belén — filamentous fungus Neurospora https://journals.biologists.com/jcs/article/115/9/1931/35045/Interaction-of-mitochondria-with-microtubules-in (old paper from 2002). Binding to motor proteins depends on peripheral proteins and ?ATP? MMM1 (outer membrane protein) not important. Also, mitochondrial “dynamics” in parasitic protists https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6871780 (2019). Many such protists have just one mito with one nucleoid — in which case mito replication is more more coupled to the cell cycle. Branched mito structures, and important of apicoplast dynamics. Asymmetry in mitochondrial structure e.g. in Toxoplasma gondii. Branches are distributed to the growing daughter cell at the last moment before cell division — cool paper also looking at dynamics of other organelles https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6810632/ . In Giardia, there are two distinct classes of mitosome (central and peripheral) separated by their physical location. Also slaved to the cell cycle. Q: are there functional correlates of these dynamics?
Iain — https://science.sciencemag.org/content/348/6232/340 human mammary stemlike cells. sometimes an asymmetric distribution of mitochondria of different ages occurs as stem cells divide. Daughters inheriting younger mitos retain stemness.
15. Roles of calcium
Belén — model of calcium transport between compartments https://febs.onlinelibrary.wiley.com/doi/full/10.1111/febs.14296 , in addition to NADH and membrane potential etc. Some coarse-grained representation of ETC activity and electrochem gradients? Size and cellular density of mitochondria are important. Roles for mitochondrial variability.
Iain PS: classic ODE models for mitochondria include Bazil https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1000632 , Beard https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.0010036 .
Also, Ca2+ signals in and out of the mitochondria, with focus on varying frequency and amplitude https://pubmed.ncbi.nlm.nih.gov/30659886/ . Roles in apoptosis. How do oscillations respond to different stimuli? Ca2+ oscillations also occur in the ER and cytosol; mitochondrial Ca2+ dynamics strongly influenced by the calcium uniporter. Ca2+ signals activate Krebs cycle enzymes and regulate cell bioenergetics. Ca2+ overload activates the permeability transition pore (mPTP) and acts as a death signal.
Apparently Ca2+ influx to mitos depends on the inter-membrane distance (volume?) — which is controlled by several players. “Low conductance mode” (of mPTP) — to allow Ca2+ regulation without triggering overload. Role for detailed shape of Ca2+ signals, in addition to frequency, in shaping cellular signals. Amplitude coding can support temperature compensation mechanisms!
Robert — Xenopus https://en.wikipedia.org/wiki/African_clawed_frog mitochondrial motility modulated by Ca2+ https://www.jneurosci.org/content/jneuro/30/10/3555.full.pdf . “during normal nerve activity, Ca elevation and activation of Na/K-ATPase act, possibly in a synergistic manner, to recruit mitochondria to a node of Ranvier to match metabolic needs.” Ca2+ enrichment slows mitochondrial motion.
[parallel question — optimal number of oDNAs per organelle?] Nucleoids studied with super-resolution microscopy https://www.sciencedirect.com/science/article/abs/pii/S1357272518302292 provide some (mammalian) observations.
Confusing claim: “Nucleoids may be attached to the inner mitochondrial membrane in fission sites, hence fragments contain always one nucleoid.”
Kostas — [optimal number of oDNAs per organelle?] — spot on survey of organelle numbers per cell https://www.frontiersin.org/articles/10.3389/fcell.2016.00085/full — metabolic needs etc. Power per mitochondrion and number of active ATP synthases https://iopscience.iop.org/article/10.1088/1478-3975/abf7d9/meta . Number of active ATP synthases can be much less than number of present ones.
ER is the main store of Ca2+ in the cell; mitochondria act as a buffer — mito uptake timescale is faster. Difference between intra and extracellular Ca2+ concentrations. Review of mitochondrial role in Ca2+ balance https://www.nature.com/articles/nrm3412 . Verbatim set of key points
- The regulation of mitochondrial Ca2+ transport in physiological and pathological conditions is controlled by channels and exchangers that are located in the outer and inner mitochondrial membrane (OMM and IMM, respectively). Whereas the OMM is permeable to solutes and ions, Ca2+ transport across the IMM is highly regulated.
- The strategic positioning of mitochondria in close proximity to Ca2+ release channels of the endoplasmic reticulum (ER) and the sarcoplasmic reticulum explains the high rate of mitochondrial Ca2+ uptake in stimulated cells, despite the low affinity of the mitochondrial Ca2+ uniporter (MCU), which mediates Ca2+ transfer through the IMM.
- The sites of contacts between mitochondria and the ER or the sarcoendoplasmic reticulum (called mitochondria associated membranes (MAMs)) are microdomains of high Ca2+ concentration. Several proteins and chaperones are responsible for the maintenance of these structures and for efficient uptake of Ca2+ released from the ER and the sarcoendoplasmic reticulum.
- Mitochondria act as intracellular Ca2+ buffers in the proximity of Ca2+ channels at the ER or the sarcoendoplasmic reticulum and the plasma membrane, thus affecting the Ca2+ feedback regulation of channel activity. Some cell types exhibit a defined distribution of mitochondria, and this affects the diffusion of Ca2+ waves through the cytosol.
- Ca2+ accumulation into the mitochondria stimulates aerobic metabolism and thus ATP production by modulating the activity of the enzymes of the tricarboxylic acid cycle (TCA cycle) and other effectors.
- Mitochondrial Ca2+ waves control cell fate. Increased levels of intracellular Ca2+ may trigger cell death by necrosis or apoptosis by causing the sustained or transient opening of a high-conductance channel of the IMM, termed the permeability transition pore (PTP). Conversely, low Ca2+ concentration levels in mitochondria cause a decrease in ATP production, which promotes pro-survival autophagy.
Also, does cell size influence chloroplast genome size? https://www.frontiersin.org/articles/10.3389/fpls.2017.02116/full
Jo — [optimal number of oDNAs per organelle?] — Frontiers paper as above. Relationship between oDNA number, organelle number, and cell requirements? Number of “virtual” mitochondria per (mammalian) cell, across different tissue types, from the 80s https://pubmed.ncbi.nlm.nih.gov/3170646/ . Scaling of organelle and cell size https://www.tandfonline.com/doi/full/10.4161/org.6.2.11464 . Iain PS: metabolic scaling, cell power and cell death https://onlinelibrary.wiley.com/doi/full/10.1002/bies.201700001 , plus “optimal cell size” from allometric scaling of mitochondria https://www.sciencedirect.com/science/article/pii/S1534580716306268
Link between Ca2+ and cell migration: fragmented mitochondria at the leading edge, more networked back at the nucleus (especially Fig 2) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5930976/ . Link between Ca2+ dynamics and mitochondrial dynamics, as mitochondria are spatially embedded buffers in moving cells.
[summer break]
16. Recent endosymbioses
Jo — Durinskia https://en.wikipedia.org/wiki/Durinskia tertiary endosymbioisis. Two different plastids? https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4217693/ https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0010711 Sequence for plastid. Are there systematic differences in gene count between primary/secondary/tertiary/etc?
Paulinella https://en.wikipedia.org/wiki/Paulinella — plastid 100mya. petL, petM absent from chromatophore. NCBI ref https://www.ncbi.nlm.nih.gov/nuccore/LC490351.1 . Some info on chromatophore genome https://www.sciencedirect.com/science/article/abs/pii/S0960982221002700
Belén — primer on endosymbiosis (and gene loss) https://www.sciencedirect.com/science/article/pii/S0960982212006550 . Paulinella “Recent work revealed that a few dozen chromatophore-derived genes have moved to the amoeba host genome, apparently some time ago since the base composition of the genes now resembles that of the host. “ — which work? Probably https://bmcbiol.biomedcentral.com/articles/10.1186/1741-7007-10-35 . Others: “The few known examples in animals include gene transfer from the reproductive parasite Wolbachia to insect and nematode host genomes, and transfer from Buchnera to the aphid nuclear genome (although the transferred Buchnera genes appear to be non-functional).“ Genome reduction in intracellular bacterial endosymbionts of insects https://www.nature.com/articles/nrmicro2670 . Protist endosymbiosies https://royalsocietypublishing.org/doi/10.1098/rstb.2009.0188 . Nested endosymbionts in mealybugs https://www.sciencedirect.com/science/article/pii/S096098221100724X ,
Iain — denitrifying organelle in ciliate 2021 https://www.ncbi.nlm.nih.gov/nuccore/LR794158.1 (paper https://www.nature.com/articles/s41586-021-03297-6 )
17. mtDNA copy number during development
Robert — horses! How mtDNA and ATP changes with embryos from young vs old mares https://repository.up.ac.za/bitstream/handle/2263/51164/Hendriks_Maternal_2015.pdf?sequence=1
3e7 mtDNA per embryo from young mares < 12y, much much less per embryo for older mares > 12y . During development, physical bottleneck https://repository.up.ac.za/bitstream/handle/2263/69199/Hendriks_Mitochondrial_2019.pdf?sequence=1
Jo — short-lived fish Nothobranchius furzeri
https://onlinelibrary.wiley.com/doi/10.1111/j.1474-9726.2011.00723.x short life, very rapid sexual maturity. Tissue-specific copy number profiles, high in energy-demanding tissues. General copy number decreases with age.
Also sea urchins https://www.cambridge.org/core/journals/zygote/article/mitochondria-during-sea-urchin-oogenesis/40B6BBA8F6D009BD73D5DC0093D515B4 — mtDNA copy num using fluorescence. Increases during oogenesis.
Kostas — Arabidopsis and Tourneria as in PLoS Biol https://onlinelibrary.wiley.com/doi/10.1111/tpj.13987 ; Arabidopsis and others https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2929101/ as in PLoS Biol. Also C elegans, roundabout route to compute mtDNA copy number [Iain — https://www.sciencedirect.com/science/article/pii/S0006291X02963941 ] also pigs https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4714324/ but this is a bit more nuanced. Also hard coral oocytes https://www.tandfonline.com/doi/pdf/10.3109/19401736.2015.1036254?needAccess=true
Belén — melon. Paternal mitochondrial transmission. In leaves, lower mtDNA count than mitochondria https://www.nature.com/articles/s41438-019-0177-8 . Subgenomic molecules — diversity in gene counts (e.g. atp vs nad). Ploidy (C count) also changes during leaf development.
18. Freestyle
Robert — https://en.wikipedia.org/wiki/Parakaryon_myojinensis Parakaryon — completely unknown position in the tree of life. Larger than prokaryotes, with (spiral) endosymbionts but no mitochondria, organelles, nucleus with only one membrane and no pores, no cytoskeleton. https://academic.oup.com/jmicro/article/61/6/423/1989140
Jo — https://en.wikipedia.org/wiki/Yareta Yareta, slow growing but very long lived flowering plant. Fluffy mats of green in arid desert. Most estimated to be over 3000 years old.
Kostas — Azolla https://en.wikipedia.org/wiki/Azolla symbiosis with nitrogen-fixing bacterium Anabaena azollae, some evidence of gene loss and transfer to nucleus? https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6786969/ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2900214/ https://microbewiki.kenyon.edu/index.php/Anabaena_azollae
Belén — https://en.wikipedia.org/wiki/Oxalis_acetosella Wood sorrel! In her garden. Very light responsive!
Iain — https://en.wikipedia.org/wiki/Ravenala Traveller’s tree, 11m long leaves, pollinated by lemurs. Also Game of Life https://en.wikipedia.org/wiki/Conway%27s_Game_of_Life
19. Newly discovered organisms
Welcome Sara and Rajneesh!
Kostas — flatworms. Kleptoplasty by some sea slugs from diatoms https://www.science.org/doi/10.1126/sciadv.aaw4337 . Endosymbiotic flatworms https://www.sciencedirect.com/science/article/pii/S0141813020334656#bb0160 retain a (previously though lost) atp8 gene
Belén — colourful orange Halloween bat https://walltrace.com/2021/02/a-new-orange-and-black-bat-species-is-always-ready-for-halloween/ discovered in abandoned mine tunnels https://digitallibrary.amnh.org/bitstream/handle/2246/7249/N3963.pdf?sequence=1&isAllowed=y
20. Model organisms
Jo — tobacco mosaic virus https://en.wikipedia.org/wiki/Tobacco_mosaic_virus . cylindrical structure with one capsomer protein. High production rate of associated protein means that we can transform in a gene of interest and produce in high amounts for production in tobacco.
Sophie — hydra https://en.wikipedia.org/wiki/Hydra_(genus) — outer and inner cell layers — not subject to senescence (death from ageing). Very simple nervous system. Tissue regeneration. Basically unlimited set of stem cells? Can reproduce clonally via budding or sexually, and seems that sexual reproduction induces ageing https://www.sciencedirect.com/science/article/pii/S0378111906005166?via%3Dihub . I see there’s also an “immortal jellyfish” https://en.wikipedia.org/wiki/Turritopsis_dohrnii
Robert — https://en.wikipedia.org/wiki/Domestic_pigeon model for navigation, homing pigeons of great use in war time, oldest domesticated bird. Map sense and compass sense — is this down to magnetoreceptors in brain? https://onlinelibrary.wiley.com/doi/10.1002/cne.10579
Recognise self in mirror. Performed better than humans at spotting humans at sea. Can distinguish between cubist and impressionist paintings. Both parents can produce milk. Also — not a model yet but interesting — https://journals.asm.org/doi/full/10.1128/mSystems.00774-20
Sara — https://en.wikipedia.org/wiki/Schizophyllum_commune . Consumption can lead to schizophrenic-like symptoms. Can contain some anti-tumour chemicals.
Rajneesh — https://en.wikipedia.org/wiki/Lotus_japonicus . Model for nitrogen fixation via microrhiza. (There’s also another model https://en.wikipedia.org/wiki/Medicago_truncatula which has different developmental setup). Short life cycle, 2-3 weeks. Small genome size.
Belén — https://en.wikipedia.org/wiki/Chlamydomonas_reinhardtii
List of more general model species it’s worth knowing:
Bacteria — E coli
Mammal — M musculus, R norvegicus
Yeast — S cerevisiae, S pombe
Invertebrate — D melanogaster, C elegans
Fish — D rerio
Angiosperm — A thaliana
Other viridiplantae — P patens, Marchantia sp.
Algae — C reinhardtii
21. Elements heavier than iron in biology
Robert — molybdenum. https://en.wikipedia.org/wiki/Molybdenum#Biological_role a 2008 research paper speculated that a scarcity of molybdenum in the Earth’s early oceans may have strongly influenced the evolution of eukaryotic life (which includes all plants and animals).[71] . Key component of nitrogenases — facilitating nitrogen fixation in cyanobacteria.
Rajneesh — plutonium (plus nitrogen and radium) isotopes and their role in ocean circulation. Plutonium isotopes — Important in tracking ocean movement and circulation. Radium isotopes in sediments and bioformation. Reminded Iain of the demon core https://en.wikipedia.org/wiki/Demon_core
Jo — iodine — the heaviest element commonly needed by life (tungsten is maybe needed in some bacterial enzymes). Important in mammalian endocrine systems, cerebrospinal fluid, arterial walls, mammary glands, etc etc. Deficiency causes goitre, swollen thyroid. We get all our iodine from food, esp seafood and plants, which in turn depend on soil. Lots of marine organisms, esp single celled, use iodine https://academic.oup.com/icb/article/49/2/155/641567 . Powerful inorganic antioxidant — particularly important in kelp https://www.pnas.org/content/105/19/6954 influencing atmosphere. Why does kelp need so much? It’s the highest accumulator of iodine. PNAS says because of oxidative exposure due to tides.
Kostas — tungsten https://en.wikipedia.org/wiki/Tungsten . first a pushback comment on the iodine paper from Jo https://academic.oup.com/icb/article/50/1/138/733337 . present in bacteria and particularly in archaea. https://www.sciencedirect.com/science/article/abs/pii/S0010854508000167 analogous role to molybdenum in some enzymes. May have been the ancestral case, since replaced by molybdenum. Some experiments attempting to exchange Mo with W. Potentially tied with the extremophilia of bacteria and archaea.
Iain — selenium — important in selenocysteine, a rare amino acid that plays a role in enzymes catalysing redox reactions. Deficiency (in China and Russia) leads to KEshan disease, but perhaps not directly — may facilitate evolution of a virus to a more dangerous form https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/keshan-disease . Too much selenium gives us selenosis https://en.wikipedia.org/wiki/Selenium#Toxicity
22. Toxicity of different elements
Arsenic
Belén – Arsenic: A compound that is a severe pollutant. Mostly found with sulfur.
It goes into the trophic chain, mainly through water, like fish and other aquatic organisms. Affects and is associated with metabolic syndromes, like diabetes. Accumulates in different tissues and fat. Useful for manufacturing pesticides. Can damage and comprise mitochondrial function -> damage energy, gene expressions, copy number. Tied with carcinogenesis. Some bacteria use arsenic in metabolism. https://pubmed.ncbi.nlm.nih.gov/20649553/
Interesting suggestion for a movie -> Arsenic and Old Lace
Rajneesh- Arsenic: High level of exposure -> neurological and heart problems
Arsenite is more dangerous compared to organic form. There are specific levels under which water and food consumption is safe (for arsenic traces). Guidelines given by WHO, but every country states its own thresholds. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4427717/#CIT0029
Kostas – Arsenic: Most of the time we get rid of it through urine. Responsible for oxidative stress. Arsenate’s structure looks like phosphate, but the latter cannot be (fully?) replaced by the former. Arsenic oxide is used in medicine for the treatment of acute promyelocytic leukemia. Arsenic compounds inhibit the PDH complex, which catalyzes the oxidation of pyruvate to acetyl-CoA by NAD+ -> cell’s energy is disrupted -> cellular apoptosis episode. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2685281/
22.5 Toxic elements (cont)
Robert – Beryllium: second lightest metal, highly charged, small so they can enter the body and accumulate in tissues and cells. Rigid, high melting point and electricity inductor. Used in industry. Can cause acute pneumonia and other diseases that look like sarcoidosis. Also causes berylliosis.
Sara – Mercury: a heavy metal, found both in inorganic and organic compounds. It’s toxic by altering mechanisms involving mitochondria, like during ATP production and causing ROS damage. Really toxic. It’s ubiquitous and volatile. Eg, found in fish. No physiological benefit by any amount of mercury (in contrast to most elements). Affects kidneys (mostly inorganic version), the nervous system (organic version).
Sophie – Lead: lead was led to the food chain (lead in bullets!) -> lead poisoning
Causes kidney failure, among others. And a main cofactor in numerous other diseases. Water contamination and poisoning. Used in cosmetics in the past.
Jo – Cadmium: Uptake by roots, common to see it near battery factories.
In https://academic.oup.com/metallomics/article/11/2/255/5957484 we see what it causes to plants. It affects all parts of the plant. Toxic in high amounts. It affects plant stomata openings. Damaging photosynthetic pathways. Competitively binds with other elements (like zinc) used by proteins. Can make cells move around and not stick to each other, which is linked with carcinogenesis. Can be nontoxic to the plant, but affects its metabolism. Poisoning from cadmium causes carcinogenesis. Affects gene expression and DNA methylation.
Kostas – Gold (and gold particles): Gold, a rare metal. Not much chemically reactive. Gold is not toxic, but ionic compounds of it (“gold salts”) are. Gold salt was used in therapies for rheumatoid arthritis, but it had some severe side effects and is responsible for specific diseases in tissues and organs.
Gold nanoparticles and nanorods. Clustered gold particles in the scale of a few nanometers. They have multiple application in medicine, like drug delivery, imaging etc.
They seem to affect mitochondria respiration and decrease mitochondrial ROS. Also, might affect the ETC. https://www.sciencedirect.com/science/article/pii/S0300483X18302452?via%3Dihub
They are probably not colocated with mitochondria, so they are less harmful than expected? https://www.sciencedirect.com/science/article/pii/S0927776509000666?via%3Dihub
Their characteristic of increasing the membrane permeability might be bad for mitochondria and leave “marks”, especially the particles with very small size.
https://pubmed.ncbi.nlm.nih.gov/19642089
23. Organisms subject to periodic environmental fluctuations
Jo – Fluctuations caused by artificial lights: Affects both mammals and plants (photorespiration) https://www.frontiersin.org/articles/10.3389/fnins.2020.602796/full
The effect on chloroplasts in https://www.intechopen.com/online-first/75498
Rajneesh – Monarch butterflies: Cosmopolitan and important in the food chain. It’s very susceptible to temperature fluxes. It applies hibernation and other defending mechanisms to protect itself. Their metabolism is tied to temperature, so if it’s too cold, they hibernate. Temperature also affects the hibernation length.
https://www.cms.int/sites/default/files/publication/fact_sheet_monarch_butterfly_climate_change.pdf
Belén – Growing plants indoors: Fluctuating environment is in favor of the plant’s life for indoors growing. Fluctuating light, temperature and humidity.
Photosynthesis and leaf condition looks like the one in the wild (natural growth).
Different scenarios to test the fluctuation intensity and periodicity (eg sinusoidal periodicity). Fast fluctuations reduced photosynthesis.
Fluctuating light produced more chlorophyll (?).Plant-specific and general traits were tested.
https://www.mdpi.com/2223-7747/9/10/1312/htm
Sara – CAM photosynthetic plants: Adapted to do photosynthesis in a “plant-hostile” environment. Open stomata during the night to keep their humidity. Pineapples are CAM plants.
Kostas – Intertidal organisms, like mussels: Most mollusks have a bunch of thread-like structures called byssels to help them attach to surfaces. They help them during tides and can alter their direction to match the sea force. A review paper about intertidal animals and how their mitochondria respond to environmental fluctuations is in https://academic.oup.com/icb/article/58/3/519/4986976
Mitochondria in these species have developed protective mechanisms to respond to sudden reoxygenation after long hypoxic exposure, by reducing proton leakage and ROS accumulation. Mitochondrial tolerance to fluctuating oxygen levels in some mollusks was associated with the high activity of mitochondrial proteases that degrade oxidatively damaged proteins thus keeping them healthy.
Also they are good with temperature and salinity fluctuations compared to other subtidal species.
More explicitly about blue mussels and how they avoid ROS formation can be found in
https://www.frontiersin.org/articles/10.3389/fmars.2021.773734/full
24. Orbiniidae with Miguel A. Meca
Belén- species Naineris Setosa as an invasive species in the Mediterranean.
There exist 13 species of native Orbiniidae in the Mediterranean. Orbiniidae have great plasticity and can become invasive species, N.Setosa is recorded as alien species in the Mediterranean area. N.Setosa was originally found in Bermuda Islands in 1900, and it is hypothesised that they occurred naturally in the Pacific Coast of America. They are widely distributed in (sub)tropical latitudes in the Western Atlantic Ocean and in tropical Pacific.
First report of N.Setosa out of its native geographic range was in an aquaculture farm in the Adriatic Sea in Italy, but it is thought that that population became extinct after the farm closed. A smaller population of N.Setosa was also found in a lagoon in Tunisia. From 2010 to 2014 samples have been taken in Santa Gilla lagoon in the southern coast of Sardinia, showing a successful establishment of the species and a stable population size. How the species arrived to this last lagoon is unknown.
N.Setosa might be associated to stressed habitats or might be a tolerant opportunistic species, as they can live in highly polluted habitats in the Mediterranean Sea or with high salinity.
https://mbr.biomedcentral.com/articles/10.1186/s41200-016-0017-6
25. Christmassy organisms
Belén — Eurasian blue tit https://en.wikipedia.org/wiki/Eurasian_blue_tit . Common around Europe; good at nesting in holes; can transmit learned behaviours including the ability to open foil-top milk bottles.
Sophie — Hazel https://en.wikipedia.org/wiki/Hazel — nuts give you wishes, and anthropogenic pollution induces dysfunctional pollen https://www.mdpi.com/1999-4907/12/1/88/htm . A note of caution about MDPI journals — not bad but sometimes viewed with suspicion
Jo — mistletoe https://en.wikipedia.org/wiki/Viscum_album — reduced mtDNA including loss of CI — why? https://www.nature.com/articles/srep17588#Abs1 . Perhaps get metabolites from host and use AOX instead? That Sci Rep paper links plastid and mito gene loss extents
Rajneesh — reindeer https://a-z-animals.com/animals/reindeer/ . Can see UV — so can see white predators. Run at 50mph. Natural heaters in nose. Famously live on lichen during snow periods.
Robert — the 12 parasites of Christmas https://www.cbsnews.com/pictures/the-12-parasites-of-christmas/ . Warble fly living on reindeer https://en.wikipedia.org/wiki/Hypoderma_tarandi. https://en.wikipedia.org/wiki/Macrophomina_phaseolina — plant pathogen fungus. Affects 500 species in 100 families including many important crops. Addressed with fungicide, or organically with solar heat traps and/or crop rotation (out of soya).
26. Christmassy organisms (2)
Sara – mistletoe see also Jo’s notes from last week (no 25). Seed dispersion using birds.
Robert – Dendrolimus sibiricus, a pine tree parasitic moth. Big outbreaks damage coniferous forests. Different developmental stages in larvae. Not easy to distinguish among species (morphological similarities) https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-015-1566-5 . Use of genetic (and mitochondrial) distances to distinguish them. Interesting genetic distribution and quantities therein.
Rajneesh – more on reindeer. Morphological changes to adapt to different and extreme environments and circumstances, like the use of anglers. Very good swimmers, as well! Only mammal to cover such long distances and see UV. Newborn reindeers can run really fast.
Kostas – Douglas-fir. Not really a fir. Leaves in moisture, slightly acidic soils. Species found in drier environments have adapted by having deeper rooting systems. Doesn’t like frost and too much cold. Low shadow-tolerance. Seed dispersion using wind and small mammals. Parasitized by different fungi, insects and even dwarf mistletoes. Differences with other related gymnosperms, especially about the size of the NDH-complex gene family and genes responsible for shade intolerance https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5592940/.
Interesting note: they show paternal cytoplasmic inheritance, maternal mitochondrial inheritance, and paternal plastid inheritance https://cdnsciencepub.com/doi/abs/10.1139/x92-010
No Responses